Gene Page: EFNB3
Summary ?
| GeneID | 1949 |
| Symbol | EFNB3 |
| Synonyms | EFL6|EPLG8|LERK8 |
| Description | ephrin-B3 |
| Reference | MIM:602297|HGNC:HGNC:3228|Ensembl:ENSG00000108947|HPRD:03804|Vega:OTTHUMG00000108161 |
| Gene type | protein-coding |
| Map location | 17p13.1 |
| Pascal p-value | 0.09 |
| Sherlock p-value | 0.489 |
| eGene | Cerebellum Meta |
| Support | Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
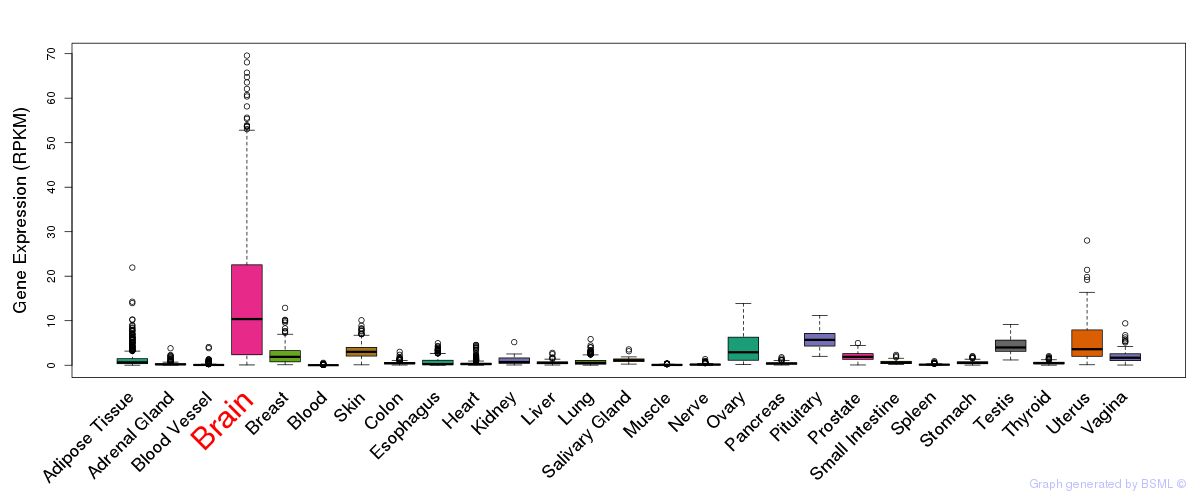
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AK3 | 0.73 | 0.67 |
| ACBD5 | 0.72 | 0.68 |
| ERLIN2 | 0.72 | 0.74 |
| KDSR | 0.71 | 0.68 |
| PIK3C2A | 0.69 | 0.70 |
| ARHGAP5 | 0.69 | 0.70 |
| GALC | 0.69 | 0.69 |
| ITGAV | 0.69 | 0.70 |
| RNF141 | 0.68 | 0.66 |
| MCCC2 | 0.68 | 0.66 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| IL32 | -0.51 | -0.46 |
| AF347015.21 | -0.44 | -0.30 |
| CLEC3B | -0.44 | -0.42 |
| FAM159B | -0.41 | -0.26 |
| AL050337.1 | -0.41 | -0.26 |
| APOC1 | -0.40 | -0.29 |
| AL353354.3 | -0.38 | -0.13 |
| GNG11 | -0.38 | -0.25 |
| AL022328.1 | -0.38 | -0.34 |
| C16orf74 | -0.38 | -0.30 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005005 | transmembrane-ephrin receptor activity | TAS | 9126477 | |
| GO:0046875 | ephrin receptor binding | IDA | 8808709 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016198 | axon choice point recognition | IEA | neuron, axon (GO term level: 14) | - |
| GO:0007399 | nervous system development | TAS | neurite (GO term level: 5) | 9126477 |
| GO:0007267 | cell-cell signaling | TAS | 9126477 | |
| GO:0007628 | adult walking behavior | IEA | - | |
| GO:0048013 | ephrin receptor signaling pathway | IDA | 8808709 | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| GO:0030154 | cell differentiation | IEA | - | |
| GO:0044419 | interspecies interaction between organisms | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016020 | membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 9126477 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG AXON GUIDANCE | 129 | 103 | All SZGR 2.0 genes in this pathway |
| PID EPHB FWD PATHWAY | 40 | 38 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA ERYTHROID DN | 493 | 298 | All SZGR 2.0 genes in this pathway |
| DOANE RESPONSE TO ANDROGEN DN | 241 | 146 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS DN | 260 | 143 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN UP | 612 | 367 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER DN | 514 | 319 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 2 UP | 30 | 24 | All SZGR 2.0 genes in this pathway |
| MANTOVANI NFKB TARGETS DN | 11 | 6 | All SZGR 2.0 genes in this pathway |
| MANTOVANI VIRAL GPCR SIGNALING DN | 49 | 37 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS DN | 357 | 212 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL FGF3 | 8 | 7 | All SZGR 2.0 genes in this pathway |
| GEISS RESPONSE TO DSRNA DN | 16 | 8 | All SZGR 2.0 genes in this pathway |
| CHIBA RESPONSE TO TSA UP | 52 | 33 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE UP | 244 | 165 | All SZGR 2.0 genes in this pathway |
| PAL PRMT5 TARGETS DN | 29 | 16 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY 4NQO OR UV | 63 | 44 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE UP | 226 | 164 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE DN | 258 | 160 | All SZGR 2.0 genes in this pathway |
| HOFFMANN PRE BI TO LARGE PRE BII LYMPHOCYTE DN | 75 | 61 | All SZGR 2.0 genes in this pathway |
| HOFFMANN LARGE TO SMALL PRE BII LYMPHOCYTE DN | 72 | 47 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3 UNMETHYLATED | 536 | 296 | All SZGR 2.0 genes in this pathway |
| NAKAYAMA SOFT TISSUE TUMORS PCA1 DN | 74 | 47 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 6 UP | 140 | 81 | All SZGR 2.0 genes in this pathway |
| CARD MIR302A TARGETS | 77 | 62 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124/506 | 921 | 927 | m8 | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-138 | 1574 | 1580 | m8 | hsa-miR-138brain | AGCUGGUGUUGUGAAUC |
| miR-145 | 913 | 920 | 1A,m8 | hsa-miR-145 | GUCCAGUUUUCCCAGGAAUCCCUU |
| miR-192/215 | 1605 | 1611 | 1A | hsa-miR-192 | CUGACCUAUGAAUUGACAGCC |
| hsa-miR-215 | AUGACCUAUGAAUUGACAGAC | ||||
| miR-204/211 | 1737 | 1744 | 1A,m8 | hsa-miR-204brain | UUCCCUUUGUCAUCCUAUGCCU |
| hsa-miR-211 | UUCCCUUUGUCAUCCUUCGCCU | ||||
| miR-28 | 1639 | 1645 | m8 | hsa-miR-28brain | AAGGAGCUCACAGUCUAUUGAG |
| miR-34/449 | 1199 | 1205 | 1A | hsa-miR-34abrain | UGGCAGUGUCUUAGCUGGUUGUU |
| hsa-miR-34c | AGGCAGUGUAGUUAGCUGAUUGC | ||||
| hsa-miR-449 | UGGCAGUGUAUUGUUAGCUGGU | ||||
| hsa-miR-449b | AGGCAGUGUAUUGUUAGCUGGC | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.