Gene Page: EPHA7
Summary ?
| GeneID | 2045 |
| Symbol | EPHA7 |
| Synonyms | EHK-3|EHK3|EK11|HEK11 |
| Description | EPH receptor A7 |
| Reference | MIM:602190|HGNC:HGNC:3390|Ensembl:ENSG00000135333|HPRD:03721|Vega:OTTHUMG00000015228 |
| Gene type | protein-coding |
| Map location | 6q16.1 |
| Pascal p-value | 0.004 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
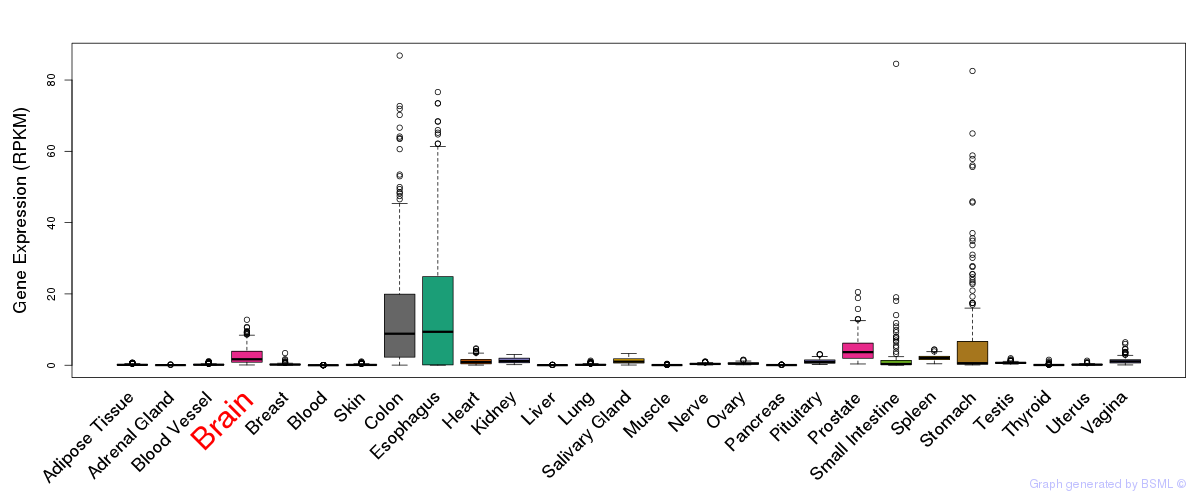
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NAPG | 0.94 | 0.94 |
| MAPK9 | 0.94 | 0.93 |
| ATP6V1C1 | 0.93 | 0.93 |
| B4GALT6 | 0.92 | 0.93 |
| IDH3A | 0.91 | 0.92 |
| GLS | 0.91 | 0.94 |
| GLRB | 0.91 | 0.92 |
| SYT1 | 0.91 | 0.90 |
| MOAP1 | 0.91 | 0.92 |
| MAP2K4 | 0.90 | 0.88 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| BCL7C | -0.49 | -0.56 |
| EFEMP2 | -0.47 | -0.48 |
| NME4 | -0.45 | -0.57 |
| CLEC3B | -0.45 | -0.52 |
| AL022328.1 | -0.45 | -0.48 |
| RAB34 | -0.44 | -0.48 |
| AC006276.2 | -0.43 | -0.43 |
| IRF7 | -0.42 | -0.53 |
| SIGIRR | -0.42 | -0.49 |
| AP003068.3 | -0.41 | -0.40 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0004872 | receptor activity | IEA | - | |
| GO:0005003 | ephrin receptor activity | IEA | - | |
| GO:0005515 | protein binding | ISS | - | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | IEA | - | |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG AXON GUIDANCE | 129 | 103 | All SZGR 2.0 genes in this pathway |
| PID EPHA FWDPATHWAY | 34 | 29 | All SZGR 2.0 genes in this pathway |
| LIU PROSTATE CANCER DN | 481 | 290 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE UP | 423 | 283 | All SZGR 2.0 genes in this pathway |
| SABATES COLORECTAL ADENOMA DN | 291 | 176 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS G UP | 238 | 135 | All SZGR 2.0 genes in this pathway |
| DAWSON METHYLATED IN LYMPHOMA TCL1 | 59 | 45 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| AFFAR YY1 TARGETS UP | 214 | 133 | All SZGR 2.0 genes in this pathway |
| JIANG AGING CEREBRAL CORTEX UP | 36 | 27 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| HILLION HMGA1B TARGETS | 92 | 68 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS UP | 165 | 118 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS DN | 138 | 92 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR DN | 505 | 328 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| KATSANOU ELAVL1 TARGETS UP | 169 | 105 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 4 | 110 | 66 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 10HR DN | 244 | 157 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 SIGNALING VIA NFIC 10HR UP | 54 | 38 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-103/107 | 73 | 79 | m8 | hsa-miR-103brain | AGCAGCAUUGUACAGGGCUAUGA |
| hsa-miR-107brain | AGCAGCAUUGUACAGGGCUAUCA | ||||
| miR-133 | 1733 | 1739 | 1A | hsa-miR-133a | UUGGUCCCCUUCAACCAGCUGU |
| hsa-miR-133b | UUGGUCCCCUUCAACCAGCUA | ||||
| miR-134 | 1940 | 1946 | m8 | hsa-miR-134brain | UGUGACUGGUUGACCAGAGGG |
| miR-137 | 1316 | 1323 | 1A,m8 | hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG |
| hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG | ||||
| miR-141/200a | 1818 | 1825 | 1A,m8 | hsa-miR-141 | UAACACUGUCUGGUAAAGAUGG |
| hsa-miR-200a | UAACACUGUCUGGUAACGAUGU | ||||
| miR-142-5p | 1335 | 1341 | 1A | hsa-miR-142-5p | CAUAAAGUAGAAAGCACUAC |
| miR-15/16/195/424/497 | 74 | 81 | 1A,m8 | hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG |
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| miR-17-5p/20/93.mr/106/519.d | 776 | 782 | m8 | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-181 | 1352 | 1358 | 1A | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-182 | 671 | 677 | m8 | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| miR-196 | 237 | 243 | m8 | hsa-miR-196a | UAGGUAGUUUCAUGUUGUUGG |
| hsa-miR-196b | UAGGUAGUUUCCUGUUGUUGG | ||||
| miR-199 | 508 | 514 | m8 | hsa-miR-199a | CCCAGUGUUCAGACUACCUGUUC |
| hsa-miR-199b | CCCAGUGUUUAGACUAUCUGUUC | ||||
| miR-203.1 | 1612 | 1618 | m8 | hsa-miR-203 | UGAAAUGUUUAGGACCACUAG |
| miR-204/211 | 177 | 184 | 1A,m8 | hsa-miR-204brain | UUCCCUUUGUCAUCCUAUGCCU |
| hsa-miR-211 | UUCCCUUUGUCAUCCUUCGCCU | ||||
| miR-26 | 144 | 150 | 1A | hsa-miR-26abrain | UUCAAGUAAUCCAGGAUAGGC |
| hsa-miR-26bSZ | UUCAAGUAAUUCAGGAUAGGUU | ||||
| miR-299-3p | 1723 | 1729 | m8 | hsa-miR-299-3p | UAUGUGGGAUGGUAAACCGCUU |
| miR-30-3p | 242 | 248 | 1A | hsa-miR-30a-3p | CUUUCAGUCGGAUGUUUGCAGC |
| hsa-miR-30e-3p | CUUUCAGUCGGAUGUUUACAGC | ||||
| miR-378* | 1031 | 1037 | 1A | hsa-miR-422b | CUGGACUUGGAGUCAGAAGGCC |
| hsa-miR-422a | CUGGACUUAGGGUCAGAAGGCC | ||||
| miR-433-3p | 347 | 353 | m8 | hsa-miR-433brain | AUCAUGAUGGGCUCCUCGGUGU |
| hsa-miR-433brain | AUCAUGAUGGGCUCCUCGGUGU | ||||
| miR-493-5p | 304 | 310 | m8 | hsa-miR-493-5p | UUGUACAUGGUAGGCUUUCAUU |
| miR-503 | 75 | 81 | 1A | hsa-miR-503 | UAGCAGCGGGAACAGUUCUGCAG |
| miR-539 | 1614 | 1620 | m8 | hsa-miR-539 | GGAGAAAUUAUCCUUGGUGUGU |
| miR-9 | 1430 | 1436 | 1A | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
| hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.