Gene Page: ESR2
Summary ?
| GeneID | 2100 |
| Symbol | ESR2 |
| Synonyms | ER-BETA|ESR-BETA|ESRB|ESTRB|Erb|NR3A2 |
| Description | estrogen receptor 2 (ER beta) |
| Reference | MIM:601663|HGNC:HGNC:3468|Ensembl:ENSG00000140009|HPRD:03390|Vega:OTTHUMG00000141306 |
| Gene type | protein-coding |
| Map location | 14q23.2 |
| Pascal p-value | 0.178 |
| TADA p-value | 0.011 |
| Fetal beta | 0.069 |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Expression | Meta-analysis of gene expression | P value: 1.461 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0521 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ESR2 | chr14 | 64735576 | G | A | NM_001040275 NM_001214902 NM_001214903 NM_001271876 NM_001271877 NM_001437 NR_073496 NR_073497 NR_073505 | p.197R>W p.197R>W p.197R>W p.197R>W p.197R>W p.197R>W . . . | missense missense missense missense missense missense npcRNA npcRNA npcRNA | Schizophrenia | DNM:Fromer_2014 |
Section II. Transcriptome annotation
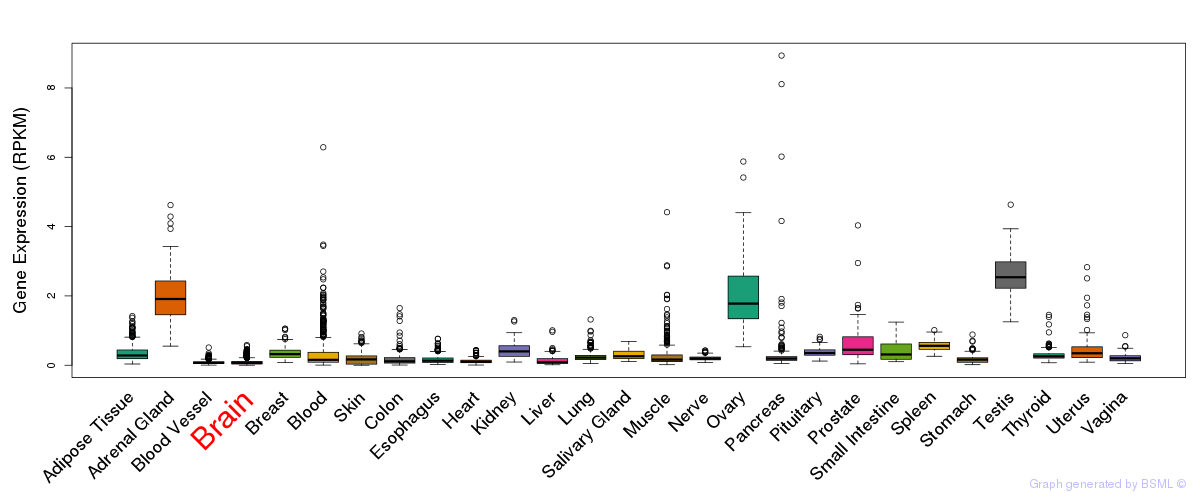
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FNDC3A | 0.78 | 0.86 |
| FAF2 | 0.76 | 0.82 |
| IMPAD1 | 0.75 | 0.83 |
| EAF1 | 0.75 | 0.82 |
| MAP3K7IP2 | 0.74 | 0.81 |
| SLC39A9 | 0.74 | 0.81 |
| MMGT1 | 0.74 | 0.80 |
| DCTN4 | 0.74 | 0.80 |
| SEC23B | 0.74 | 0.78 |
| ZFR | 0.73 | 0.80 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FXYD1 | -0.59 | -0.62 |
| MT-CO2 | -0.56 | -0.60 |
| AF347015.21 | -0.55 | -0.60 |
| HIGD1B | -0.55 | -0.60 |
| CSAG1 | -0.54 | -0.57 |
| AC098691.2 | -0.54 | -0.62 |
| IFI27 | -0.53 | -0.57 |
| AF347015.8 | -0.53 | -0.58 |
| AF347015.31 | -0.52 | -0.58 |
| AF347015.33 | -0.52 | -0.55 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003700 | transcription factor activity | TAS | 8769313 | |
| GO:0003713 | transcription coactivator activity | TAS | 8769313 | |
| GO:0005496 | steroid binding | TAS | 11181953 | |
| GO:0005515 | protein binding | IPI | 12039952 |14764652 | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0048019 | receptor antagonist activity | NAS | 9671811 | |
| GO:0030284 | estrogen receptor activity | TAS | 11181953 | |
| GO:0046872 | metal ion binding | IEA | - | |
| GO:0043565 | sequence-specific DNA binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006355 | regulation of transcription, DNA-dependent | TAS | 11181953 | |
| GO:0030520 | estrogen receptor signaling pathway | TAS | 11181953 | |
| GO:0006350 | transcription | IEA | - | |
| GO:0007267 | cell-cell signaling | TAS | 8769313 | |
| GO:0007165 | signal transduction | TAS | 8769313 |10749889 | |
| GO:0030308 | negative regulation of cell growth | NAS | 11181953 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | TAS | 11181953 | |
| GO:0005739 | mitochondrion | IDA | 15024130 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BCAS2 | DAM1 | breast carcinoma amplified sequence 2 | BCAS2 interacts with ER-beta. This interaction was modeled on a demonstrated interaction between human BCAS2 and ER-beta from an unspecified species. | BIND | 15694360 |
| CALM1 | CALML2 | CAMI | DD132 | PHKD | calmodulin 1 (phosphorylase kinase, delta) | - | HPRD | 11981030 |
| CALM2 | CAMII | PHKD | PHKD2 | calmodulin 2 (phosphorylase kinase, delta) | - | HPRD | 11981030 |
| CALM3 | PHKD | PHKD3 | calmodulin 3 (phosphorylase kinase, delta) | - | HPRD | 11981030 |
| DAP3 | DAP-3 | DKFZp686G12159 | MGC126058 | MGC126059 | MRP-S29 | MRPS29 | bMRP-10 | death associated protein 3 | Reconstituted Complex | BioGRID | 10903152 |
| DDX54 | DP97 | MGC2835 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 54 | - | HPRD,BioGRID | 12466272 |
| DNTTIP2 | ERBP | FCF2 | HSU15552 | LPTS-RP2 | MGC163494 | RP4-561L24.1 | TdIF2 | deoxynucleotidyltransferase, terminal, interacting protein 2 | - | HPRD,BioGRID | 15047147 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | ER alpha interacts with ER beta to form heterodimer. This interaction was modeled on a demonstrated interaction between human ER alpha and mouse ER beta. | BIND | 10706629 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | - | HPRD,BioGRID | 9473491 |
| MAD2L1 | HSMAD2 | MAD2 | MAD2 mitotic arrest deficient-like 1 (yeast) | MAD2 interacts with ER beta. This interaction was modeled on a demonstrated interaction between MAD2 from human and sheep and ER beta from rat and mouse. | BIND | 10706629 |
| MAD2L1 | HSMAD2 | MAD2 | MAD2 mitotic arrest deficient-like 1 (yeast) | - | HPRD,BioGRID | 10706629 |
| MED1 | CRSP1 | CRSP200 | DRIP205 | DRIP230 | MGC71488 | PBP | PPARBP | PPARGBP | RB18A | TRAP220 | TRIP2 | mediator complex subunit 1 | - | HPRD,BioGRID | 11867769 |
| MKNK2 | GPRK7 | MNK2 | MAP kinase interacting serine/threonine kinase 2 | - | HPRD,BioGRID | 11013076 |
| MMS19 | FLJ34167 | FLJ95146 | MET18 | MGC99604 | MMS19L | hMMS19 | MMS19 nucleotide excision repair homolog (S. cerevisiae) | - | HPRD | 11279242 |
| NCOA1 | F-SRC-1 | KAT13A | MGC129719 | MGC129720 | NCoA-1 | RIP160 | SRC-1 | SRC1 | bHLHe42 | nuclear receptor coactivator 1 | - | HPRD,BioGRID | 10809226 |
| NCOA3 | ACTR | AIB-1 | AIB1 | CAGH16 | CTG26 | KAT13B | MGC141848 | RAC3 | SRC3 | TNRC14 | TNRC16 | TRAM-1 | pCIP | nuclear receptor coactivator 3 | - | HPRD,BioGRID | 10681591 |11389589 |
| NCOA6 | AIB3 | ANTP | ASC2 | HOX1.1 | HOXA7 | KIAA0181 | NRC | PRIP | RAP250 | TRBP | nuclear receptor coactivator 6 | - | HPRD,BioGRID | 11773444 |
| NR0B2 | FLJ17090 | SHP | SHP1 | nuclear receptor subfamily 0, group B, member 2 | - | HPRD,BioGRID | 9773978 |10648597 |
| OXT | MGC126890 | MGC126892 | OT | OT-NPI | oxytocin, prepropeptide | - | HPRD | 9501254 |
| PELP1 | HMX3 | MNAR | P160 | proline, glutamate and leucine rich protein 1 | - | HPRD,BioGRID | 12415108 |
| PIAS2 | MGC102682 | MIZ1 | PIASX | PIASX-ALPHA | PIASX-BETA | SIZ2 | ZMIZ4 | miz | protein inhibitor of activated STAT, 2 | Reconstituted Complex | BioGRID | 11117529 |
| PRMT2 | HRMT1L1 | MGC111373 | protein arginine methyltransferase 2 | PRMT2 interacts with ER-beta. | BIND | 12039952 |
| PSMC3IP | GT198 | HOP2 | HUMGT198A | TBPIP | PSMC3 interacting protein | - | HPRD | 11739747 |
| RBM39 | CAPER | CAPERalpha | CC1.3 | DKFZp781C0423 | FLJ44170 | HCC1 | RNPC2 | fSAP59 | RNA binding motif protein 39 | - | HPRD,BioGRID | 11704680 |
| RBM9 | FOX2 | Fox-2 | HNRBP2 | HRNBP2 | RTA | dJ106I20.3 | fxh | RNA binding motif protein 9 | - | HPRD,BioGRID | 11875103 |
| REXO4 | REX4 | XPMC2 | XPMC2H | REX4, RNA exonuclease 4 homolog (S. cerevisiae) | - | HPRD,BioGRID | 10908561 |
| SMARCE1 | BAF57 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 | - | HPRD,BioGRID | 12145209 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | Affinity Capture-Western | BioGRID | 11032808 |
| STAT5A | MGF | STAT5 | signal transducer and activator of transcription 5A | - | HPRD,BioGRID | 11682624 |
| TRIM24 | PTC6 | RNF82 | TF1A | TIF1 | TIF1A | TIF1ALPHA | hTIF1 | tripartite motif-containing 24 | - | HPRD | 10598587 |
| YWHAH | YWHA1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide | - | HPRD | 11266503 |
| YWHAQ | 14-3-3 | 1C5 | HS1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide | Reconstituted Complex | BioGRID | 11266503 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID ER NONGENOMIC PATHWAY | 41 | 35 | All SZGR 2.0 genes in this pathway |
| PID ERB GENOMIC PATHWAY | 15 | 8 | All SZGR 2.0 genes in this pathway |
| PID ERA GENOMIC PATHWAY | 65 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME GENERIC TRANSCRIPTION PATHWAY | 352 | 181 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | 49 | 36 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| LI CYTIDINE ANALOGS CYCTOTOXICITY | 15 | 8 | All SZGR 2.0 genes in this pathway |
| LOPES METHYLATED IN COLON CANCER UP | 27 | 20 | All SZGR 2.0 genes in this pathway |
| HILLION HMGA1 TARGETS | 90 | 71 | All SZGR 2.0 genes in this pathway |
| MATZUK PREOVULATORY FOLLICLE | 10 | 8 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 ICP WITH H3K4ME3 AND H3K27ME3 | 34 | 21 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS ICP WITH H3K4ME3 AND H327ME3 | 126 | 83 | All SZGR 2.0 genes in this pathway |
| LI PROSTATE CANCER EPIGENETIC | 30 | 22 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS UP | 221 | 135 | All SZGR 2.0 genes in this pathway |
| SENGUPTA EBNA1 ANTICORRELATED | 173 | 85 | All SZGR 2.0 genes in this pathway |