Gene Page: F2
Summary ?
| GeneID | 2147 |
| Symbol | F2 |
| Synonyms | PT|RPRGL2|THPH1 |
| Description | coagulation factor II, thrombin |
| Reference | MIM:176930|HGNC:HGNC:3535|Ensembl:ENSG00000180210|HPRD:01488|Vega:OTTHUMG00000150344 |
| Gene type | protein-coding |
| Map location | 11p11 |
| Pascal p-value | 8.276E-8 |
| eGene | Cerebellum |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias,schizotypal | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
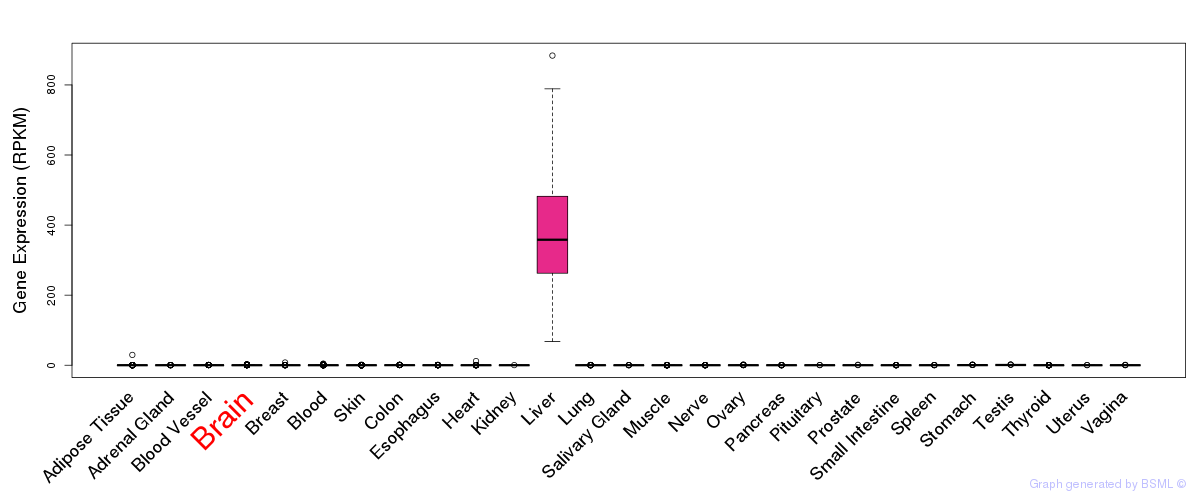
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| LYVE1 | 0.82 | 0.84 |
| SIGLEC1 | 0.81 | 0.78 |
| LILRB5 | 0.79 | 0.79 |
| CD163 | 0.78 | 0.85 |
| DAB2 | 0.78 | 0.70 |
| SLC6A20 | 0.74 | 0.70 |
| CD209 | 0.73 | 0.76 |
| MPEG1 | 0.72 | 0.62 |
| CYP1B1 | 0.70 | 0.85 |
| KIAA1199 | 0.69 | 0.79 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SNHG12 | -0.34 | -0.48 |
| RPS27A | -0.32 | -0.56 |
| RPS21 | -0.32 | -0.54 |
| RPL23A | -0.32 | -0.53 |
| RPL37 | -0.32 | -0.55 |
| NME4 | -0.31 | -0.46 |
| RPL34 | -0.31 | -0.55 |
| RPL31 | -0.30 | -0.51 |
| RPS20 | -0.30 | -0.54 |
| RPS15A | -0.30 | -0.51 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005102 | receptor binding | IPI | Neurotransmitter (GO term level: 4) | 12855810 |
| GO:0004252 | serine-type endopeptidase activity | IDA | glutamate (GO term level: 7) | 12855810 |
| GO:0005509 | calcium ion binding | IEA | - | |
| GO:0008233 | peptidase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006508 | proteolysis | TAS | 7507931 | |
| GO:0007262 | STAT protein nuclear translocation | TAS | 10692450 | |
| GO:0007260 | tyrosine phosphorylation of STAT protein | TAS | 10692450 | |
| GO:0007166 | cell surface receptor linked signal transduction | IDA | 1672265 | |
| GO:0030168 | platelet activation | TAS | 12855810 | |
| GO:0009611 | response to wounding | TAS | 12855810 | |
| GO:0006953 | acute-phase response | IEA | - | |
| GO:0006919 | caspase activation | TAS | 10692450 | |
| GO:0006915 | apoptosis | TAS | 10692450 | |
| GO:0007275 | multicellular organismal development | TAS | 9639571 | |
| GO:0030194 | positive regulation of blood coagulation | IDA | 9038223 | |
| GO:0042730 | fibrinolysis | IDA | 12855810 | |
| GO:0051209 | release of sequestered calcium ion into cytosol | IDA | 1672265 | |
| GO:0032967 | positive regulation of collagen biosynthetic process | IDA | 9639571 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | EXP | 2538457 |2742826 |2866798 |12907439 |14982929 | |
| GO:0005576 | extracellular region | NAS | 14718574 | |
| GO:0005615 | extracellular space | TAS | 3242619 | |
| GO:0005625 | soluble fraction | TAS | 2719946 | |
| GO:0005886 | plasma membrane | EXP | 2538457 |12907439 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| C6 | - | complement component 6 | - | HPRD | 3192535 |
| CPB2 | CPU | PCPB | TAFI | carboxypeptidase B2 (plasma) | Reconstituted Complex | BioGRID | 8663147 |
| F11 | FXI | MGC141891 | coagulation factor XI | - | HPRD | 1652157 |
| F2R | CF2R | HTR | PAR1 | TR | coagulation factor II (thrombin) receptor | Thrombin interacts with PAR1. This interaction was modeled on a demonstrated interaction between Thrombin from an unspecified species and human PAR1. | BIND | 15707890 |
| F2RL2 | PAR3 | coagulation factor II (thrombin) receptor-like 2 | - | HPRD | 12732212 |
| F2RL3 | PAR4 | coagulation factor II (thrombin) receptor-like 3 | - | HPRD | 12732212 |
| F5 | FVL | PCCF | coagulation factor V (proaccelerin, labile factor) | - | HPRD,BioGRID | 9032460 |
| F9 | FIX | HEMB | MGC129641 | MGC129642 | PTC | coagulation factor IX | - | HPRD | 15039440 |
| FGA | Fib2 | MGC119422 | MGC119423 | MGC119425 | fibrinogen alpha chain | - | HPRD,BioGRID | 1587268 |2133223 |
| GP1BA | BSS | CD42B | CD42b-alpha | GP1B | MGC34595 | glycoprotein Ib (platelet), alpha polypeptide | - | HPRD | 2933256 |
| ITGA2B | CD41 | CD41B | GP2B | GPIIb | GTA | HPA3 | integrin, alpha 2b (platelet glycoprotein IIb of IIb/IIIa complex, antigen CD41) | - | HPRD,BioGRID | 11487023 |
| ITGA5 | CD49e | FNRA | VLA5A | integrin, alpha 5 (fibronectin receptor, alpha polypeptide) | - | HPRD | 9864377 |
| KNG1 | BDK | KNG | kininogen 1 | - | HPRD | 1652157 |
| PLAT | DKFZp686I03148 | T-PA | TPA | plasminogen activator, tissue | Co-crystal Structure | BioGRID | 10543954 |
| SERPINA5 | PAI3 | PCI | PLANH3 | PROCI | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 | - | HPRD | 12878585 |
| SERPINB6 | CAP | DKFZp686I04222 | MGC111370 | MSTP057 | PI6 | PTI | SPI3 | serpin peptidase inhibitor, clade B (ovalbumin), member 6 | Reconstituted Complex | BioGRID | 8415716 |
| SERPINB8 | CAP2 | PI8 | serpin peptidase inhibitor, clade B (ovalbumin), member 8 | - | HPRD | 9402754 |
| SERPINC1 | AT3 | ATIII | MGC22579 | serpin peptidase inhibitor, clade C (antithrombin), member 1 | - | HPRD | 3800906 |11927130 |
| SERPIND1 | D22S673 | HC2 | HCF2 | HCII | HLS2 | LS2 | serpin peptidase inhibitor, clade D (heparin cofactor), member 1 | Co-crystal Structure | BioGRID | 12169660 |
| SERPIND1 | D22S673 | HC2 | HCF2 | HCII | HLS2 | LS2 | serpin peptidase inhibitor, clade D (heparin cofactor), member 1 | - | HPRD | 2760054 |
| SERPINE1 | PAI | PAI-1 | PAI1 | PLANH1 | serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 | Reconstituted Complex | BioGRID | 12709053 |
| SERPING1 | C1IN | C1INH | C1NH | HAE1 | HAE2 | serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 | - | HPRD,BioGRID | 11460008 |
| THBD | CD141 | THRM | TM | thrombomodulin | - | HPRD | 7615164 |8663147 |
| THBD | CD141 | THRM | TM | thrombomodulin | Reconstituted Complex | BioGRID | 2544585 |8663147 |
| THBS1 | THBS | TSP | TSP1 | thrombospondin 1 | - | HPRD,BioGRID | 2435757 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| KEGG COMPLEMENT AND COAGULATION CASCADES | 69 | 49 | All SZGR 2.0 genes in this pathway |
| KEGG REGULATION OF ACTIN CYTOSKELETON | 216 | 144 | All SZGR 2.0 genes in this pathway |
| BIOCARTA AMI PATHWAY | 20 | 13 | All SZGR 2.0 genes in this pathway |
| BIOCARTA SPPA PATHWAY | 22 | 18 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BIOPEPTIDES PATHWAY | 45 | 35 | All SZGR 2.0 genes in this pathway |
| BIOCARTA EXTRINSIC PATHWAY | 13 | 9 | All SZGR 2.0 genes in this pathway |
| BIOCARTA FIBRINOLYSIS PATHWAY | 12 | 8 | All SZGR 2.0 genes in this pathway |
| BIOCARTA INTRINSIC PATHWAY | 23 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NFAT PATHWAY | 56 | 45 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PLATELETAPP PATHWAY | 14 | 10 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PAR1 PATHWAY | 37 | 28 | All SZGR 2.0 genes in this pathway |
| ST GA12 PATHWAY | 23 | 16 | All SZGR 2.0 genes in this pathway |
| PID ANGIOPOIETIN RECEPTOR PATHWAY | 50 | 41 | All SZGR 2.0 genes in this pathway |
| PID HNF3B PATHWAY | 45 | 37 | All SZGR 2.0 genes in this pathway |
| PID THROMBIN PAR4 PATHWAY | 15 | 11 | All SZGR 2.0 genes in this pathway |
| PID SYNDECAN 4 PATHWAY | 32 | 25 | All SZGR 2.0 genes in this pathway |
| PID THROMBIN PAR1 PATHWAY | 43 | 32 | All SZGR 2.0 genes in this pathway |
| REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | 27 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | 11 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | 91 | 65 | All SZGR 2.0 genes in this pathway |
| REACTOME GASTRIN CREB SIGNALLING PATHWAY VIA PKC AND MAPK | 205 | 136 | All SZGR 2.0 genes in this pathway |
| REACTOME COMMON PATHWAY | 14 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME PEPTIDE LIGAND BINDING RECEPTORS | 188 | 108 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | 305 | 177 | All SZGR 2.0 genes in this pathway |
| REACTOME DIABETES PATHWAYS | 133 | 91 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | 16 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF PROTEINS | 518 | 242 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA Q SIGNALLING EVENTS | 184 | 116 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | 32 | 22 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR LIGAND BINDING | 408 | 246 | All SZGR 2.0 genes in this pathway |
| REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | 32 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME POST TRANSLATIONAL PROTEIN MODIFICATION | 188 | 116 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET AGGREGATION PLUG FORMATION | 36 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME INTRINSIC PATHWAY | 17 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | 208 | 138 | All SZGR 2.0 genes in this pathway |
| ROVERSI GLIOMA COPY NUMBER UP | 100 | 75 | All SZGR 2.0 genes in this pathway |
| CERVERA SDHB TARGETS 1 DN | 38 | 22 | All SZGR 2.0 genes in this pathway |
| HSIAO LIVER SPECIFIC GENES | 244 | 154 | All SZGR 2.0 genes in this pathway |
| LEE AGING CEREBELLUM UP | 84 | 58 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS DN | 138 | 92 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS DN | 366 | 257 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH INV 16 TRANSLOCATION | 422 | 277 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 9 11 TRANSLOCATION | 130 | 87 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS LCP WITH H3K4ME3 | 174 | 100 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S3 | 266 | 180 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT DN | 222 | 141 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES LCP WITH H3K4ME3 | 142 | 80 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
| NABA ECM REGULATORS | 238 | 125 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |