Gene Page: ABCD1
Summary ?
| GeneID | 215 |
| Symbol | ABCD1 |
| Synonyms | ABC42|ALD|ALDP|AMN |
| Description | ATP binding cassette subfamily D member 1 |
| Reference | MIM:300371|HGNC:HGNC:61|Ensembl:ENSG00000101986|HPRD:02300|Vega:OTTHUMG00000024215 |
| Gene type | protein-coding |
| Map location | Xq28 |
| Fetal beta | 0.121 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10994209 | chr10 | 61877705 | ABCD1 | 215 | 0.07 | trans |
Section II. Transcriptome annotation
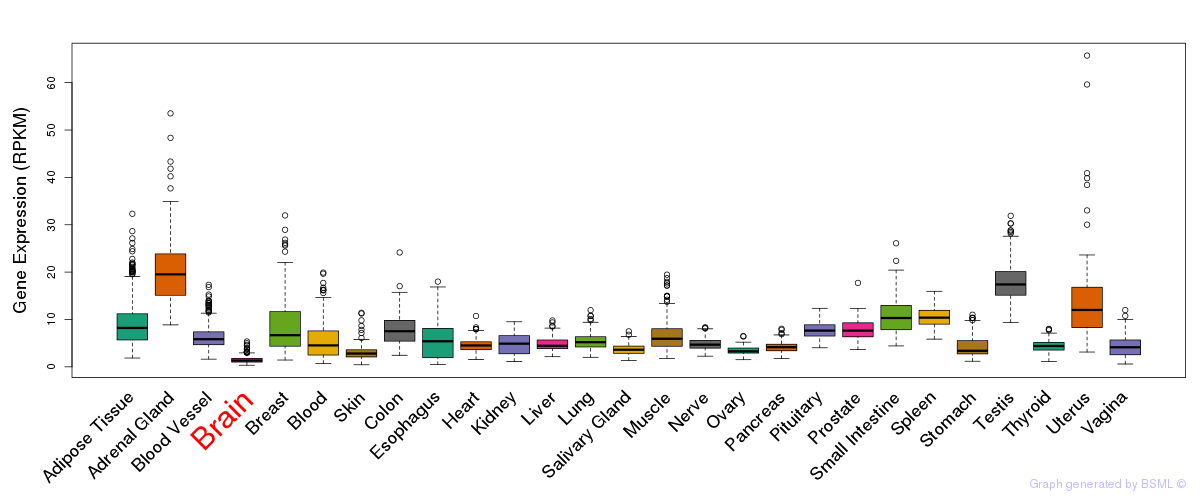
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FOLR1 | 0.61 | 0.26 |
| CP | 0.59 | 0.33 |
| C2orf40 | 0.58 | 0.34 |
| KCNJ13 | 0.57 | 0.21 |
| CLIC6 | 0.56 | 0.31 |
| TTR | 0.56 | 0.09 |
| CLDN3 | 0.55 | 0.29 |
| ABCA4 | 0.54 | 0.17 |
| RBM47 | 0.53 | 0.21 |
| TYRP1 | 0.53 | 0.45 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| DACT1 | -0.21 | 0.01 |
| KLHL1 | -0.21 | -0.01 |
| SSBP2 | -0.20 | -0.01 |
| PCSK9 | -0.20 | -0.09 |
| SCUBE1 | -0.20 | -0.20 |
| SRGAP1 | -0.20 | -0.07 |
| CBLB | -0.20 | -0.01 |
| SEMA3A | -0.20 | -0.02 |
| SLA | -0.20 | 0.03 |
| SH3BP2 | -0.19 | -0.21 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ABC TRANSPORTERS | 44 | 29 | All SZGR 2.0 genes in this pathway |
| KEGG PEROXISOME | 78 | 47 | All SZGR 2.0 genes in this pathway |
| REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | 18 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | 12 | 6 | All SZGR 2.0 genes in this pathway |
| REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | 34 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMEMBRANE TRANSPORT OF SMALL MOLECULES | 413 | 270 | All SZGR 2.0 genes in this pathway |
| REACTOME PEROXISOMAL LIPID METABOLISM | 21 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 MUTATED SIGNATURE 1 DN | 126 | 86 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 SIGNATURE 3 DN | 162 | 116 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA UP | 544 | 308 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| JAZAG TGFB1 SIGNALING UP | 108 | 69 | All SZGR 2.0 genes in this pathway |
| JAZAG TGFB1 SIGNALING VIA SMAD4 DN | 66 | 38 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS IMPRINTED AND X LINKED | 17 | 8 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS DN | 411 | 249 | All SZGR 2.0 genes in this pathway |
| SHIPP DLBCL CURED VS FATAL DN | 45 | 30 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS DN | 366 | 238 | All SZGR 2.0 genes in this pathway |
| SU PANCREAS | 54 | 30 | All SZGR 2.0 genes in this pathway |
| HOFFMANN IMMATURE TO MATURE B LYMPHOCYTE DN | 50 | 36 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS UP | 395 | 249 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |