Gene Page: FGA
Summary ?
| GeneID | 2243 |
| Symbol | FGA |
| Synonyms | Fib2 |
| Description | fibrinogen alpha chain |
| Reference | MIM:134820|HGNC:HGNC:3661|Ensembl:ENSG00000171560|HPRD:00619|Vega:OTTHUMG00000150330 |
| Gene type | protein-coding |
| Map location | 4q28 |
| Pascal p-value | 0.667 |
| Fetal beta | 0.3 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
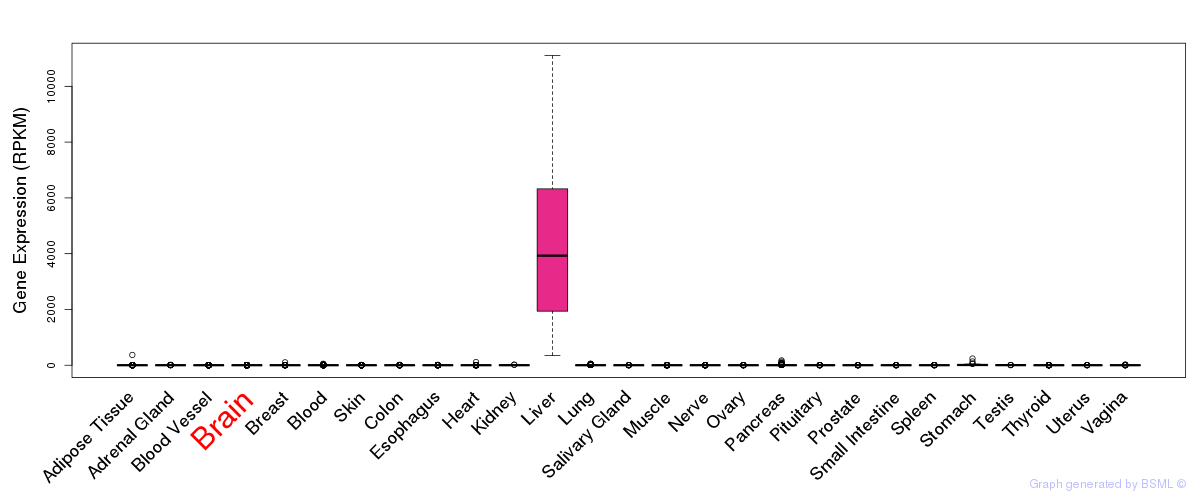
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GRB2 | 0.86 | 0.89 |
| CCDC109A | 0.84 | 0.89 |
| ARL8A | 0.83 | 0.85 |
| XPNPEP1 | 0.83 | 0.86 |
| AASDHPPT | 0.82 | 0.85 |
| RAB11A | 0.82 | 0.86 |
| PNMA1 | 0.82 | 0.83 |
| YWHAZ | 0.82 | 0.84 |
| RAB1A | 0.82 | 0.84 |
| ARPC2 | 0.82 | 0.85 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.72 | -0.72 |
| AF347015.2 | -0.70 | -0.73 |
| AF347015.33 | -0.70 | -0.72 |
| AF347015.31 | -0.70 | -0.72 |
| AF347015.8 | -0.69 | -0.71 |
| MT-CYB | -0.69 | -0.70 |
| FXYD1 | -0.68 | -0.71 |
| AF347015.26 | -0.68 | -0.71 |
| AF347015.27 | -0.68 | -0.71 |
| AF347015.15 | -0.67 | -0.69 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005102 | receptor binding | IEA | Neurotransmitter (GO term level: 4) | - |
| GO:0030674 | protein binding, bridging | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007165 | signal transduction | IEA | - | |
| GO:0030168 | platelet activation | IEA | - | |
| GO:0051258 | protein polymerization | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | EXP | 2742826 |5084810 |7356959 |10605720 | |
| GO:0005576 | extracellular region | NAS | 14718574 | |
| GO:0005577 | fibrinogen complex | IEA | - | |
| GO:0005577 | fibrinogen complex | TAS | 10467729 | |
| GO:0005886 | plasma membrane | EXP | 10605720 | |
| GO:0031093 | platelet alpha granule lumen | EXP | 5084810 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BAT2 | D6S51 | D6S51E | DKFZp686D09175 | G2 | HLA-B associated transcript 2 | - | HPRD,BioGRID | 14667819 |
| CDH5 | 7B4 | CD144 | FLJ17376 | cadherin 5, type 2 (vascular endothelium) | - | HPRD,BioGRID | 11900554 |
| F13A1 | F13A | coagulation factor XIII, A1 polypeptide | - | HPRD,BioGRID | 10956659 |
| F2 | PT | coagulation factor II (thrombin) | - | HPRD,BioGRID | 1587268 |2133223 |
| FGA | Fib2 | MGC119422 | MGC119423 | MGC119425 | fibrinogen alpha chain | - | HPRD,BioGRID | 9689040 |
| FGB | MGC104327 | MGC120405 | fibrinogen beta chain | - | HPRD,BioGRID | 12356313 |
| FGG | - | fibrinogen gamma chain | - | HPRD,BioGRID | 12356313 |
| HRG | DKFZp779H1622 | HPRG | HRGP | histidine-rich glycoprotein | - | HPRD,BioGRID | 9276466 |
| ITGA2B | CD41 | CD41B | GP2B | GPIIb | GTA | HPA3 | integrin, alpha 2b (platelet glycoprotein IIb of IIb/IIIa complex, antigen CD41) | - | HPRD | 11460491 |
| ITGB3 | CD61 | GP3A | GPIIIa | integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) | - | HPRD | 11460491 |
| NID1 | NID | nidogen 1 | - | HPRD,BioGRID | 1680863 |
| PLAT | DKFZp686I03148 | T-PA | TPA | plasminogen activator, tissue | - | HPRD,BioGRID | 3088041 |11170397 |
| SERPINA5 | PAI3 | PCI | PLANH3 | PROCI | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 | - | HPRD,BioGRID | 8384496 |8589203 |
| SERPINF2 | A2AP | AAP | ALPHA-2-PI | API | PLI | serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2 | - | HPRD | 10963790 |14751930 |
| THBS1 | THBS | TSP | TSP1 | thrombospondin 1 | - | HPRD,BioGRID | 9867861 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG COMPLEMENT AND COAGULATION CASCADES | 69 | 49 | All SZGR 2.0 genes in this pathway |
| BIOCARTA AMI PATHWAY | 20 | 13 | All SZGR 2.0 genes in this pathway |
| BIOCARTA EXTRINSIC PATHWAY | 13 | 9 | All SZGR 2.0 genes in this pathway |
| BIOCARTA FIBRINOLYSIS PATHWAY | 12 | 8 | All SZGR 2.0 genes in this pathway |
| BIOCARTA INTRINSIC PATHWAY | 23 | 15 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN1 PATHWAY | 66 | 44 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN3 PATHWAY | 43 | 25 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN2 PATHWAY | 29 | 18 | All SZGR 2.0 genes in this pathway |
| PID UPA UPAR PATHWAY | 42 | 30 | All SZGR 2.0 genes in this pathway |
| PID EPHRINB REV PATHWAY | 30 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | 89 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRIN CELL SURFACE INTERACTIONS | 79 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME COMMON PATHWAY | 14 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | 15 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | 15 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | 27 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | 32 | 22 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET AGGREGATION PLUG FORMATION | 36 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME AMYLOIDS | 83 | 63 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | 208 | 138 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA UP | 544 | 308 | All SZGR 2.0 genes in this pathway |
| CAVARD LIVER CANCER MALIGNANT VS BENIGN | 32 | 19 | All SZGR 2.0 genes in this pathway |
| DAUER STAT3 TARGETS UP | 49 | 35 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER DN | 514 | 319 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS DN | 357 | 212 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB MORPHOLINO DN | 200 | 112 | All SZGR 2.0 genes in this pathway |
| HSIAO LIVER SPECIFIC GENES | 244 | 154 | All SZGR 2.0 genes in this pathway |
| KAAB FAILED HEART ATRIUM UP | 38 | 30 | All SZGR 2.0 genes in this pathway |
| ZHANG ANTIVIRAL RESPONSE TO RIBAVIRIN UP | 30 | 21 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D DN | 270 | 181 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G123 DN | 51 | 30 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT DN | 222 | 141 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE G2 M | 216 | 124 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR DN | 88 | 59 | All SZGR 2.0 genes in this pathway |
| LU EZH2 TARGETS UP | 295 | 155 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 DN | 448 | 282 | All SZGR 2.0 genes in this pathway |
| ALFANO MYC TARGETS | 239 | 156 | All SZGR 2.0 genes in this pathway |
| NABA ECM GLYCOPROTEINS | 196 | 99 | All SZGR 2.0 genes in this pathway |
| NABA CORE MATRISOME | 275 | 148 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-101 | 128 | 135 | 1A,m8 | hsa-miR-101 | UACAGUACUGUGAUAACUGAAG |
| miR-144 | 129 | 135 | 1A | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| miR-29 | 107 | 113 | 1A | hsa-miR-29aSZ | UAGCACCAUCUGAAAUCGGUU |
| hsa-miR-29bSZ | UAGCACCAUUUGAAAUCAGUGUU | ||||
| hsa-miR-29cSZ | UAGCACCAUUUGAAAUCGGU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.