Gene Page: KDM1A
Summary ?
| GeneID | 23028 |
| Symbol | KDM1A |
| Synonyms | AOF2|BHC110|CPRF|KDM1|LSD1 |
| Description | lysine demethylase 1A |
| Reference | MIM:609132|HGNC:HGNC:29079|Ensembl:ENSG00000004487|HPRD:09800|Vega:OTTHUMG00000003220 |
| Gene type | protein-coding |
| Map location | 1p36.12 |
| Pascal p-value | 0.465 |
| Sherlock p-value | 0.317 |
| Fetal beta | 1.795 |
| eGene | Myers' cis & trans |
| Support | Ascano FMRP targets Chromatin Remodeling Genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs4854166 | chr2 | 3297005 | KDM1A | 23028 | 0.18 | trans | ||
| rs6902183 | chr6 | 121963786 | KDM1A | 23028 | 0.15 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
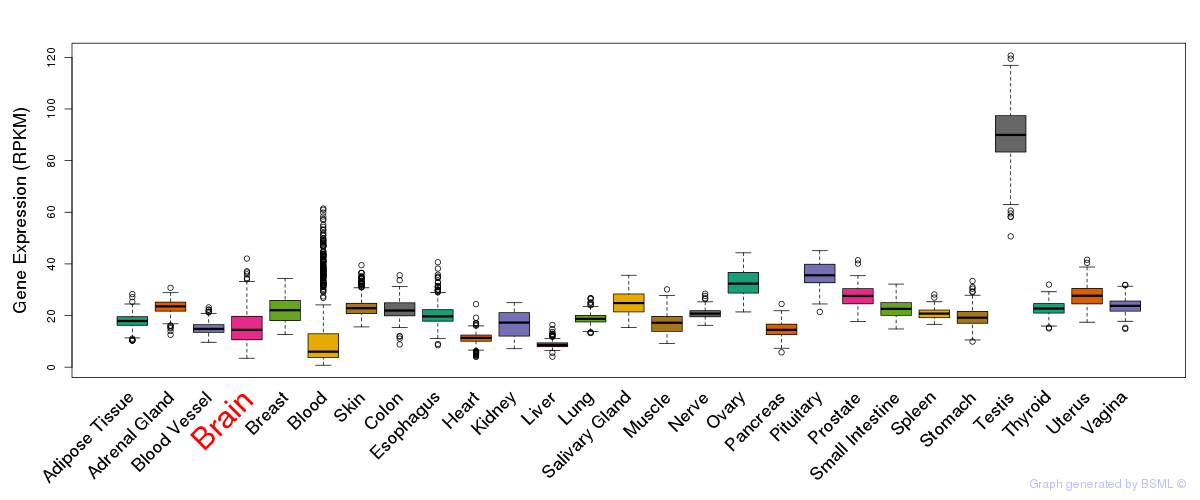
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AOF2 | BHC110 | KDM1 | KIAA0601 | LSD1 | amine oxidase (flavin containing) domain 2 | Affinity Capture-MS | BioGRID | 12493763 |
| ASCC2 | ASC1p100 | activating signal cointegrator 1 complex subunit 2 | Two-hybrid | BioGRID | 16169070 |
| ATP5J2 | ATP5JL | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F2 | Two-hybrid | BioGRID | 16169070 |
| CCDC90B | MDS011 | MDS025 | MGC104239 | coiled-coil domain containing 90B | Two-hybrid | BioGRID | 16169070 |
| GDF9 | - | growth differentiation factor 9 | Two-hybrid | BioGRID | 16169070 |
| GTF2I | BAP-135 | BAP135 | BTKAP1 | DIWS | FLJ38776 | FLJ56355 | IB291 | SPIN | TFII-I | WBS | WBSCR6 | general transcription factor II, i | Affinity Capture-MS Affinity Capture-Western | BioGRID | 12493763 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | - | HPRD,BioGRID | 12493763 |
| HDAC2 | RPD3 | YAF1 | histone deacetylase 2 | Affinity Capture-MS Affinity Capture-Western Co-purification | BioGRID | 9804427 |12032298 |12493763 |
| HIST3H3 | H3.4 | H3/g | H3FT | H3t | MGC126886 | MGC126888 | histone cluster 3, H3 | LSD1 interacts with and demethylates H3. | BIND | 15620353 |
| HMG20B | BRAF25 | BRAF35 | FLJ26127 | HMGX2 | PP7706 | SMARCE1r | SOXL | pp8857 | high-mobility group 20B | Affinity Capture-MS Affinity Capture-Western Co-purification | BioGRID | 12032298 |12493763 |
| IMMT | DKFZp779P1653 | HMP | MGC111146 | P87 | P87/89 | P89 | PIG4 | PIG52 | inner membrane protein, mitochondrial (mitofilin) | Two-hybrid | BioGRID | 16169070 |
| KIAA0182 | GSE1 | KIAA0182 | Affinity Capture-MS | BioGRID | 12493763 |
| PHF21A | BHC80 | BM-006 | KIAA1696 | PHD finger protein 21A | - | HPRD,BioGRID | 15325272 |
| RCOR1 | COREST | KIAA0071 | RCOR | REST corepressor 1 | Affinity Capture-MS Affinity Capture-Western Co-purification | BioGRID | 11171972 |12032298 |
| SETDB1 | ESET | KG1T | KIAA0067 | KMT1E | SET domain, bifurcated 1 | Two-hybrid | BioGRID | 16169070 |
| SIN3A | DKFZp434K2235 | FLJ90319 | KIAA0700 | SIN3 homolog A, transcription regulator (yeast) | Affinity Capture-Western | BioGRID | 11171972 |
| UNC119 | HRG4 | unc-119 homolog (C. elegans) | Two-hybrid | BioGRID | 16169070 |
| ZMYM2 | FIM | MYM | RAMP | SCLL | ZNF198 | zinc finger, MYM-type 2 | Affinity Capture-MS | BioGRID | 12493763 |
| ZMYM3 | DXS6673E | KIAA0385 | MYM | XFIM | ZNF198L2 | ZNF261 | zinc finger, MYM-type 3 | Affinity Capture-MS | BioGRID | 12493763 |
| ZNF217 | ZABC1 | zinc finger protein 217 | Affinity Capture-MS | BioGRID | 12493763 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID NOTCH PATHWAY | 59 | 49 | All SZGR 2.0 genes in this pathway |
| PID AR PATHWAY | 61 | 46 | All SZGR 2.0 genes in this pathway |
| PID HES HEY PATHWAY | 48 | 39 | All SZGR 2.0 genes in this pathway |
| REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | 132 | 101 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| BORCZUK MALIGNANT MESOTHELIOMA UP | 305 | 185 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| HATADA METHYLATED IN LUNG CANCER UP | 390 | 236 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA2 PCC NETWORK | 423 | 265 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION DN | 517 | 309 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES CORE NINE CORRELATED | 100 | 68 | All SZGR 2.0 genes in this pathway |
| ROZANOV MMP14 TARGETS UP | 266 | 171 | All SZGR 2.0 genes in this pathway |
| PENG LEUCINE DEPRIVATION DN | 187 | 122 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| HEDENFALK BREAST CANCER HEREDITARY VS SPORADIC | 50 | 32 | All SZGR 2.0 genes in this pathway |
| HEDENFALK BREAST CANCER BRCA1 VS BRCA2 | 163 | 113 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RESPONSE TO TRABECTEDIN DN | 271 | 175 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| KONDO PROSTATE CANCER HCP WITH H3K27ME3 | 97 | 72 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER DN | 238 | 145 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA PRONEURAL | 177 | 132 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE EARLY LATE | 317 | 190 | All SZGR 2.0 genes in this pathway |
| FOSTER KDM1A TARGETS DN | 211 | 119 | All SZGR 2.0 genes in this pathway |