Gene Page: ARHGEF9
Summary ?
| GeneID | 23229 |
| Symbol | ARHGEF9 |
| Synonyms | COLLYBISTIN|EIEE8|HPEM-2|PEM-2|PEM2 |
| Description | Cdc42 guanine nucleotide exchange factor 9 |
| Reference | MIM:300429|HGNC:HGNC:14561|Ensembl:ENSG00000131089|HPRD:02337|Vega:OTTHUMG00000021700 |
| Gene type | protein-coding |
| Map location | Xq11.1 |
| Sherlock p-value | 0.976 |
| Fetal beta | -1.156 |
| Support | G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.humanPSD G2Cdb.humanPSP Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
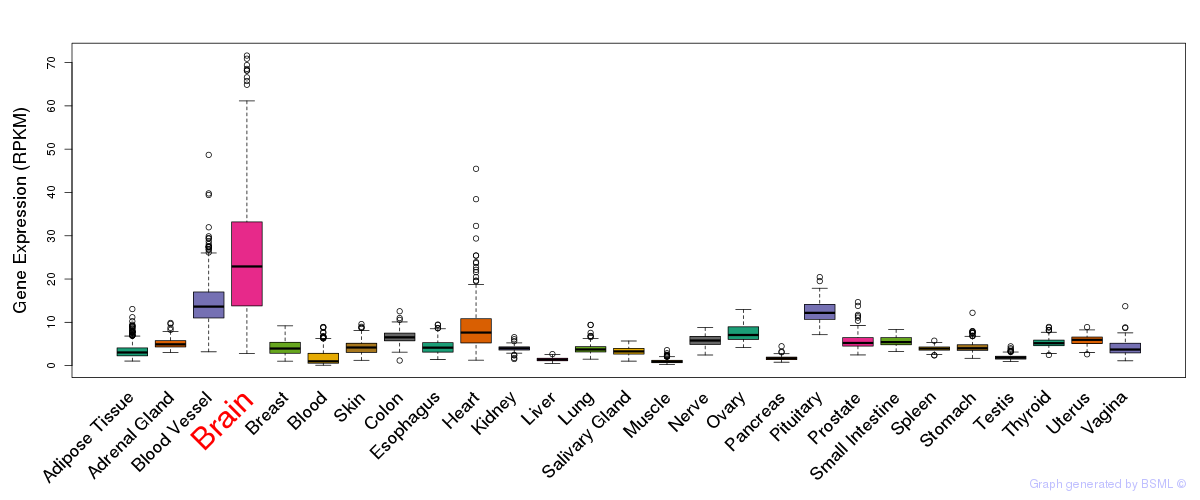
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| APRT | 0.85 | 0.83 |
| SSR4 | 0.85 | 0.82 |
| SF3B5 | 0.85 | 0.82 |
| FAM96B | 0.85 | 0.84 |
| ATP5G2 | 0.85 | 0.80 |
| COPE | 0.84 | 0.85 |
| AIP | 0.84 | 0.80 |
| C22orf32 | 0.83 | 0.81 |
| PSMB4 | 0.83 | 0.81 |
| EIF3K | 0.83 | 0.80 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| COBLL1 | -0.44 | -0.47 |
| Z83840.4 | -0.42 | -0.47 |
| AHNAK | -0.41 | -0.49 |
| GIMAP8 | -0.41 | -0.45 |
| PTRF | -0.40 | -0.43 |
| FLT1 | -0.40 | -0.47 |
| ABCB1 | -0.40 | -0.43 |
| MT-ATP8 | -0.40 | -0.46 |
| AF347015.26 | -0.40 | -0.46 |
| EPAS1 | -0.39 | -0.43 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA MYOSIN PATHWAY | 31 | 22 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PAR1 PATHWAY | 37 | 28 | All SZGR 2.0 genes in this pathway |
| PID CDC42 REG PATHWAY | 30 | 22 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY RHO GTPASES | 113 | 81 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | 186 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME NRAGE SIGNALS DEATH THROUGH JNK | 43 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | 60 | 43 | All SZGR 2.0 genes in this pathway |
| REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | 81 | 61 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME NEUROTRANSMITTER RECEPTOR BINDING AND DOWNSTREAM TRANSMISSION IN THE POSTSYNAPTIC CELL | 137 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMEMBRANE TRANSPORT OF SMALL MOLECULES | 413 | 270 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA1213 SIGNALLING EVENTS | 74 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME GABA A RECEPTOR ACTIVATION | 12 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME GABA RECEPTOR ACTIVATION | 52 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME ION CHANNEL TRANSPORT | 55 | 42 | All SZGR 2.0 genes in this pathway |
| REACTOME LIGAND GATED ION CHANNEL TRANSPORT | 21 | 21 | All SZGR 2.0 genes in this pathway |
| ZHONG RESPONSE TO AZACITIDINE AND TSA DN | 70 | 38 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST NORMAL DUCTAL VS LOBULAR DN | 7 | 6 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE UP | 423 | 283 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER BASAL VS LULMINAL | 330 | 215 | All SZGR 2.0 genes in this pathway |
| DAIRKEE TERT TARGETS UP | 380 | 213 | All SZGR 2.0 genes in this pathway |
| YANG BREAST CANCER ESR1 DN | 25 | 15 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 24HR DN | 148 | 102 | All SZGR 2.0 genes in this pathway |
| BILD HRAS ONCOGENIC SIGNATURE | 261 | 166 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 | 718 | 401 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC ICP WITH H3K4ME3 | 445 | 257 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA PRONEURAL | 177 | 132 | All SZGR 2.0 genes in this pathway |