Gene Page: CLASP1
Summary ?
| GeneID | 23332 |
| Symbol | CLASP1 |
| Synonyms | MAST1 |
| Description | cytoplasmic linker associated protein 1 |
| Reference | MIM:605852|HGNC:HGNC:17088|Ensembl:ENSG00000074054|HPRD:09322|Vega:OTTHUMG00000153331 |
| Gene type | protein-coding |
| Map location | 2q14.2-q14.3 |
| Pascal p-value | 0.139 |
| Sherlock p-value | 0.882 |
| Fetal beta | 0.909 |
| DMG | 2 (# studies) |
| Support | CompositeSet Darnell FMRP targets Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 3 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 3 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.023 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00755 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg20340149 | 2 | 122407145 | CLASP1 | 1.066E-4 | -0.209 | 0.028 | DMG:Wockner_2014 |
| cg14574905 | 2 | 122176293 | CLASP1 | 3.483E-4 | 0.358 | 0.041 | DMG:Wockner_2014 |
| cg01297674 | 2 | 122406765 | CLASP1 | 3.03E-9 | -0.017 | 2.09E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
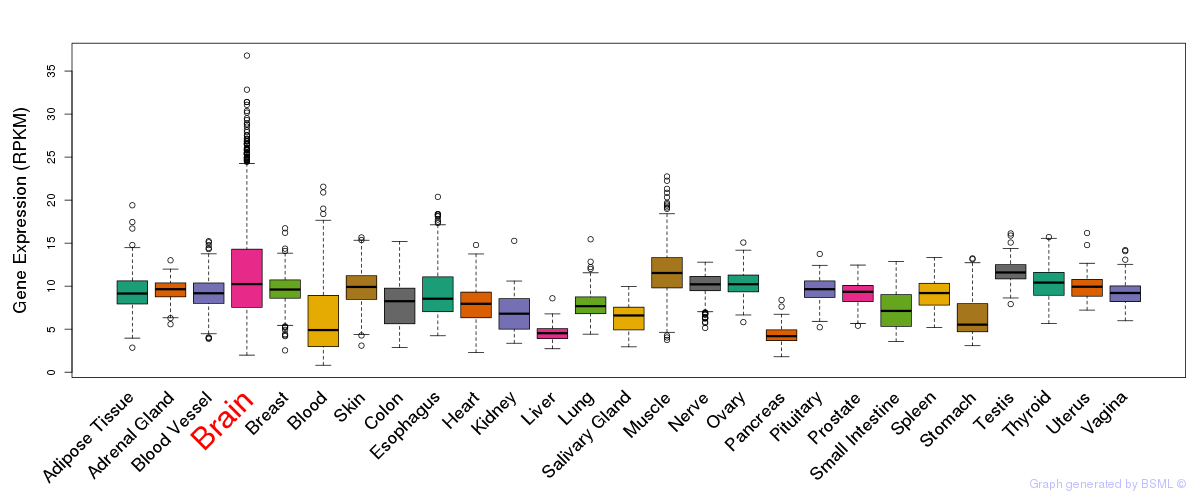
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SNX17 | 0.87 | 0.90 |
| C20orf29 | 0.87 | 0.86 |
| RPUSD3 | 0.86 | 0.84 |
| ACTR1B | 0.86 | 0.85 |
| RHBDD2 | 0.85 | 0.88 |
| UBL7 | 0.85 | 0.86 |
| ABHD14A | 0.85 | 0.84 |
| TRUB2 | 0.84 | 0.87 |
| P4HTM | 0.84 | 0.84 |
| C20orf30 | 0.84 | 0.87 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| EIF5B | -0.57 | -0.64 |
| AC010300.1 | -0.49 | -0.57 |
| AC005921.3 | -0.48 | -0.66 |
| AF347015.18 | -0.47 | -0.32 |
| NSBP1 | -0.45 | -0.42 |
| ANP32C | -0.43 | -0.45 |
| ZNF326 | -0.39 | -0.35 |
| AC100783.1 | -0.39 | -0.31 |
| HSP90AB4P | -0.38 | -0.42 |
| SFRS18 | -0.38 | -0.52 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 15631994 |17342765 | |
| GO:0051010 | microtubule plus-end binding | IDA | 12837247 | |
| GO:0043515 | kinetochore binding | IMP | 12837247 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001578 | microtubule bundle formation | IMP | 12837247 | |
| GO:0007026 | negative regulation of microtubule depolymerization | IGI | 15631994 | |
| GO:0007026 | negative regulation of microtubule depolymerization | IMP | 16866869 | |
| GO:0007163 | establishment or maintenance of cell polarity | NAS | 15928712 | |
| GO:0007049 | cell cycle | IEA | - | |
| GO:0007067 | mitosis | IEA | - | |
| GO:0010458 | exit from mitosis | IMP | 16866869 | |
| GO:0051301 | cell division | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000777 | condensed chromosome kinetochore | IEA | - | |
| GO:0005794 | Golgi apparatus | IDA | 15631994 | |
| GO:0005828 | kinetochore microtubule | TAS | 12837247 | |
| GO:0005856 | cytoskeleton | IEA | - | |
| GO:0005881 | cytoplasmic microtubule | IDA | 11290329 | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0030981 | cortical microtubule cytoskeleton | IDA | 12837247 | |
| GO:0031592 | centrosomal corona | IDA | 16914514 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | 66 | 43 | All SZGR 2.0 genes in this pathway |
| REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | 59 | 38 | All SZGR 2.0 genes in this pathway |
| REACTOME AXON GUIDANCE | 251 | 188 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ROBO RECEPTOR | 30 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC M M G1 PHASES | 172 | 98 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC G2 G2 M PHASES | 81 | 50 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA REPLICATION | 192 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC PROMETAPHASE | 87 | 51 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA UP | 294 | 178 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| LUI THYROID CANCER PAX8 PPARG UP | 44 | 29 | All SZGR 2.0 genes in this pathway |
| LUI THYROID CANCER CLUSTER 1 | 51 | 33 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS EARLY PROGENITOR | 532 | 309 | All SZGR 2.0 genes in this pathway |
| ULE SPLICING VIA NOVA2 | 43 | 38 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK DN | 318 | 220 | All SZGR 2.0 genes in this pathway |
| DAZARD UV RESPONSE CLUSTER G6 | 153 | 112 | All SZGR 2.0 genes in this pathway |
| OUILLETTE CLL 13Q14 DELETION UP | 74 | 40 | All SZGR 2.0 genes in this pathway |
| CHENG IMPRINTED BY ESTRADIOL | 110 | 68 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 9 11 TRANSLOCATION | 130 | 87 | All SZGR 2.0 genes in this pathway |
| WINNEPENNINCKX MELANOMA METASTASIS UP | 162 | 86 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| LINSLEY MIR16 TARGETS | 206 | 127 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-135 | 229 | 235 | 1A | hsa-miR-135a | UAUGGCUUUUUAUUCCUAUGUGA |
| hsa-miR-135b | UAUGGCUUUUCAUUCCUAUGUG | ||||
| miR-181 | 161 | 168 | 1A,m8 | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-194 | 2918 | 2924 | 1A | hsa-miR-194 | UGUAACAGCAACUCCAUGUGGA |
| miR-200bc/429 | 115 | 121 | m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-214 | 256 | 263 | 1A,m8 | hsa-miR-214brain | ACAGCAGGCACAGACAGGCAG |
| miR-219 | 110 | 116 | 1A | hsa-miR-219brain | UGAUUGUCCAAACGCAAUUCU |
| miR-320 | 133 | 139 | m8 | hsa-miR-320 | AAAAGCUGGGUUGAGAGGGCGAA |
| miR-323 | 2999 | 3005 | m8 | hsa-miR-323brain | GCACAUUACACGGUCGACCUCU |
| miR-370 | 1844 | 1850 | 1A | hsa-miR-370brain | GCCUGCUGGGGUGGAACCUGG |
| miR-493-5p | 2978 | 2984 | m8 | hsa-miR-493-5p | UUGUACAUGGUAGGCUUUCAUU |
| miR-494 | 2950 | 2956 | 1A | hsa-miR-494brain | UGAAACAUACACGGGAAACCUCUU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.