Gene Page: TDRD7
Summary ?
| GeneID | 23424 |
| Symbol | TDRD7 |
| Synonyms | CATC4|PCTAIRE2BP|TRAP |
| Description | tudor domain containing 7 |
| Reference | MIM:611258|HGNC:HGNC:30831|Ensembl:ENSG00000196116|HPRD:11626|Vega:OTTHUMG00000020326 |
| Gene type | protein-coding |
| Map location | 9q22.33 |
| Pascal p-value | 0.77 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg24339935 | 9 | 100174143 | TDRD7 | 5.9E-5 | -0.211 | 0.023 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1561519 | chr2 | 35823020 | TDRD7 | 23424 | 0.19 | trans | ||
| rs7591339 | chr2 | 35825597 | TDRD7 | 23424 | 0.19 | trans | ||
| rs1371418 | chr2 | 35830333 | TDRD7 | 23424 | 0.11 | trans | ||
| rs7570146 | chr2 | 35844751 | TDRD7 | 23424 | 0.19 | trans | ||
| rs1439691 | chr2 | 35851281 | TDRD7 | 23424 | 0.19 | trans |
Section II. Transcriptome annotation
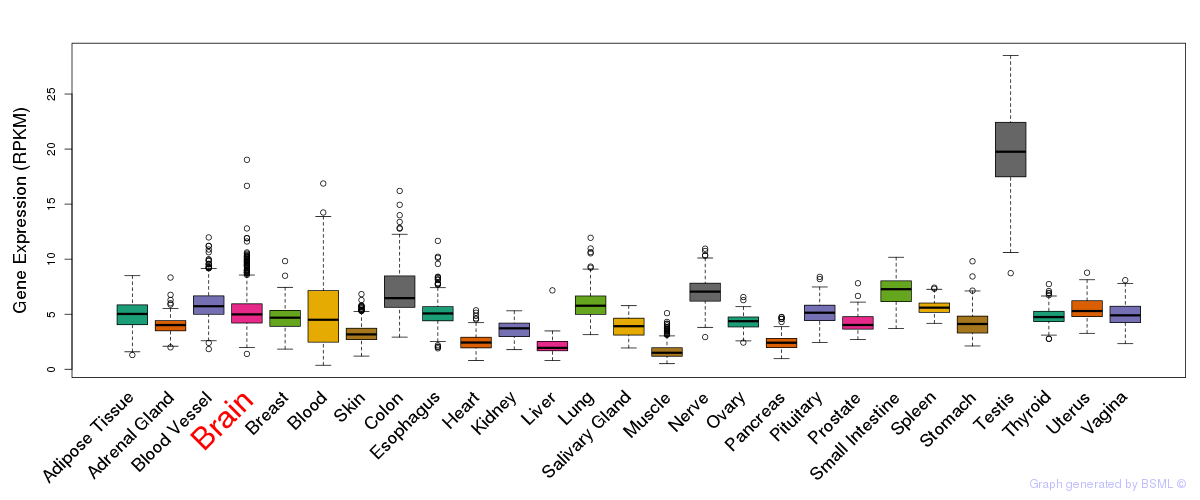
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| LRFN5 | 0.73 | 0.80 |
| PNMA5 | 0.70 | 0.76 |
| P4HA2 | 0.69 | 0.78 |
| CMAS | 0.69 | 0.78 |
| LRRC4C | 0.68 | 0.77 |
| YWHAH | 0.68 | 0.78 |
| PANK2 | 0.68 | 0.75 |
| OPTN | 0.67 | 0.72 |
| P2RX5 | 0.67 | 0.77 |
| ARMCX2 | 0.66 | 0.71 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| IMPA2 | -0.43 | -0.50 |
| AF347015.21 | -0.42 | -0.26 |
| AF347015.2 | -0.42 | -0.28 |
| AF347015.18 | -0.42 | -0.31 |
| AF347015.8 | -0.41 | -0.28 |
| MT-CO2 | -0.40 | -0.27 |
| AF347015.26 | -0.40 | -0.28 |
| PLA2G5 | -0.39 | -0.24 |
| EMX2 | -0.38 | -0.50 |
| MT-CYB | -0.38 | -0.26 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID AURORA A PATHWAY | 31 | 20 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 DN | 267 | 178 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 UP | 380 | 236 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 UP | 418 | 263 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| DAUER STAT3 TARGETS DN | 50 | 34 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| ROSS AML WITH MLL FUSIONS | 78 | 45 | All SZGR 2.0 genes in this pathway |
| BENNETT SYSTEMIC LUPUS ERYTHEMATOSUS | 31 | 21 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION UP | 314 | 201 | All SZGR 2.0 genes in this pathway |
| BROWNE INTERFERON RESPONSIVE GENES | 68 | 44 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| ZHENG GLIOBLASTOMA PLASTICITY UP | 250 | 168 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA D DN | 78 | 34 | All SZGR 2.0 genes in this pathway |
| VALK AML CLUSTER 16 | 26 | 17 | All SZGR 2.0 genes in this pathway |
| JISON SICKLE CELL DISEASE UP | 181 | 106 | All SZGR 2.0 genes in this pathway |
| ZEMBUTSU SENSITIVITY TO CYCLOPHOSPHAMIDE | 18 | 12 | All SZGR 2.0 genes in this pathway |
| BOSCO INTERFERON INDUCED ANTIVIRAL MODULE | 78 | 48 | All SZGR 2.0 genes in this pathway |