Gene Page: MTOR
Summary ?
| GeneID | 2475 |
| Symbol | MTOR |
| Synonyms | FRAP|FRAP1|FRAP2|RAFT1|RAPT1|SKS |
| Description | mechanistic target of rapamycin |
| Reference | MIM:601231|HGNC:HGNC:3942|Ensembl:ENSG00000198793|HPRD:03134|Vega:OTTHUMG00000002001 |
| Gene type | protein-coding |
| Map location | 1p36.2 |
| Pascal p-value | 0.882 |
| Sherlock p-value | 0.316 |
| Support | CompositeSet Darnell FMRP targets Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Xu_2012 | Whole Exome Sequencing analysis | De novo mutations of 4 genes were identified by exome sequencing of 795 samples in this study | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| MTOR | chr1 | 11293489 | T | C | NM_004958 | p.796N>S | missense | Schizophrenia | DNM:Xu_2012 |
Section II. Transcriptome annotation
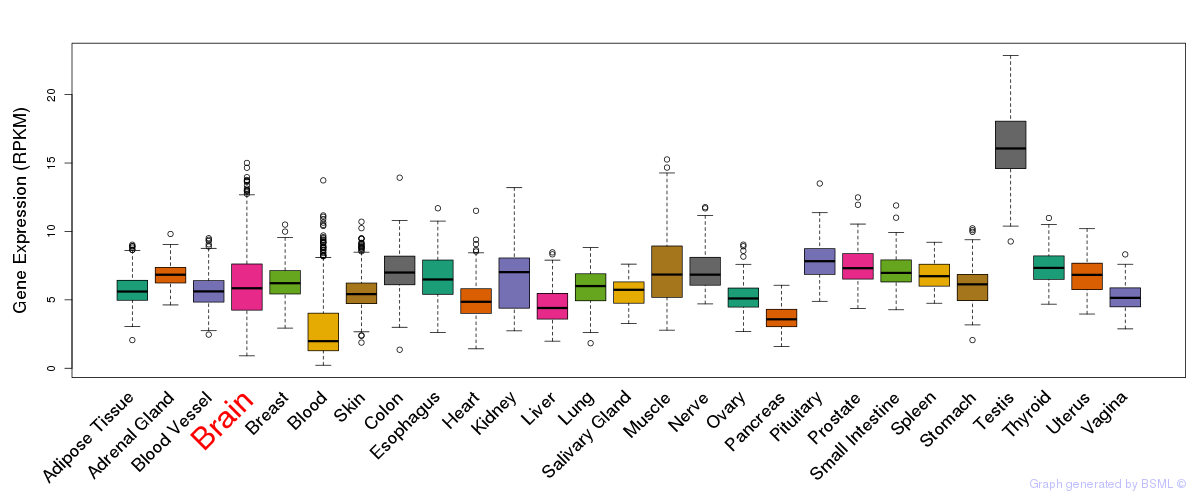
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| BRCA2 | 0.95 | 0.77 |
| MCM10 | 0.95 | 0.76 |
| KIF14 | 0.94 | 0.74 |
| TOP2A | 0.94 | 0.80 |
| TIMELESS | 0.94 | 0.78 |
| CENPF | 0.94 | 0.82 |
| KIF15 | 0.94 | 0.78 |
| NCAPH | 0.94 | 0.75 |
| HELLS | 0.94 | 0.78 |
| ASPM | 0.94 | 0.78 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HLA-F | -0.46 | -0.70 |
| C5orf53 | -0.46 | -0.69 |
| FBXO2 | -0.46 | -0.66 |
| LHPP | -0.45 | -0.61 |
| PTH1R | -0.45 | -0.67 |
| AIFM3 | -0.44 | -0.68 |
| ALDOC | -0.44 | -0.65 |
| ASPHD1 | -0.44 | -0.57 |
| TNFSF12 | -0.43 | -0.65 |
| AF347015.27 | -0.43 | -0.74 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ABL1 | ABL | JTK7 | bcr/abl | c-ABL | p150 | v-abl | c-abl oncogene 1, receptor tyrosine kinase | Affinity Capture-Western | BioGRID | 10753870 |
| AKT1 | AKT | MGC99656 | PKB | PKB-ALPHA | PRKBA | RAC | RAC-ALPHA | v-akt murine thymoma viral oncogene homolog 1 | Protein-peptide Reconstituted Complex | BioGRID | 10910062 |14970221 |15718470 |
| CFP | BFD | PFC | PFD | PROPERDIN | complement factor properdin | Reconstituted Complex | BioGRID | 11733037 |
| CLIP1 | CLIP | CLIP-170 | CLIP170 | CYLN1 | MGC131604 | RSN | CAP-GLY domain containing linker protein 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 12231510 |
| EIF3B | EIF3-ETA | EIF3-P110 | EIF3-P116 | EIF3S9 | MGC104664 | MGC131875 | PRT1 | eukaryotic translation initiation factor 3, subunit B | Affinity Capture-Western | BioGRID | 16286006 |
| EIF3C | EIF3S8 | FLJ78287 | MGC189744 | eIF3-p110 | eukaryotic translation initiation factor 3, subunit C | Affinity Capture-Western | BioGRID | 16286006 |
| EIF3F | EIF3S5 | eIF3-p47 | eukaryotic translation initiation factor 3, subunit F | Affinity Capture-Western Two-hybrid | BioGRID | 16541103 |
| EIF4EBP1 | 4E-BP1 | 4EBP1 | BP-1 | MGC4316 | PHAS-I | eukaryotic translation initiation factor 4E binding protein 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 9465032 |10971657 |12150925 |12150926 |12747827 |15767663 |15854902 |16798736 |
| FKBP1A | FKBP-12 | FKBP1 | FKBP12 | FKBP12C | PKC12 | PKCI2 | PPIASE | FK506 binding protein 1A, 12kDa | - | HPRD,BioGRID | 8662507 |
| FKBP1A | FKBP-12 | FKBP1 | FKBP12 | FKBP12C | PKC12 | PKCI2 | PPIASE | FK506 binding protein 1A, 12kDa | mTOR interacts with FKBP12 bound to rapamycin. This interaction was modeled on a demonstrated interaction between human mTOR and mouse FKBP12. | BIND | 15467718 |
| FRAP1 | FLJ44809 | FRAP | FRAP2 | MTOR | RAFT1 | RAPT1 | FK506 binding protein 12-rapamycin associated protein 1 | Affinity Capture-Western | BioGRID | 16870609 |17360675 |
| GBL | LST8 | MGC111011 | POP3 | WAT1 | G protein beta subunit-like | Affinity Capture-MS Affinity Capture-Western | BioGRID | 12719976 |15066126 |16183647 |
| GDI2P | - | GDP dissociation inhibitor 2 pseudogene | Affinity Capture-Western | BioGRID | 11287630 |
| GPHN | GEPH | GPH | GPHRYN | KIAA1385 | gephyrin | - | HPRD,BioGRID | 10325225 |
| IRS1 | HIRS-1 | insulin receptor substrate 1 | Affinity Capture-Western | BioGRID | 11287630 |
| KIAA1303 | - | raptor | - | HPRD,BioGRID | 12150925 |
| KIAA1303 | - | raptor | mTOR interacts with raptor. | BIND | 15718470 |
| LRPAP1 | A2MRAP | A2RAP | HBP44 | MGC138272 | MRAP | RAP | low density lipoprotein receptor-related protein associated protein 1 | Affinity Capture-Western | BioGRID | 16798736 |
| MAPKAP1 | JC310 | MGC2745 | MIP1 | SIN1 | SIN1b | SIN1g | mitogen-activated protein kinase associated protein 1 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 16962653 |17043309 |
| PA2G4 | EBP1 | HG4-1 | p38-2G4 | proliferation-associated 2G4, 38kDa | Reconstituted Complex | BioGRID | 15896331 |
| PDK1 | - | pyruvate dehydrogenase kinase, isozyme 1 | Phenotypic Enhancement | BioGRID | 10567431 |
| PLD2 | - | phospholipase D2 | Affinity Capture-Western | BioGRID | 16837165 |
| PPP2R2A | B55-ALPHA | B55A | FLJ26613 | MGC52248 | PR52A | PR55A | protein phosphatase 2 (formerly 2A), regulatory subunit B, alpha isoform | Reconstituted Complex | BioGRID | 10200280 |
| PRKAA1 | AMPK | AMPKa1 | MGC33776 | MGC57364 | protein kinase, AMP-activated, alpha 1 catalytic subunit | Protein-peptide | BioGRID | 14970221 |
| PRKCD | MAY1 | MGC49908 | PKCD | nPKC-delta | protein kinase C, delta | Affinity Capture-Western | BioGRID | 10698949 |
| RAP1A | KREV-1 | KREV1 | RAP1 | SMGP21 | RAP1A, member of RAS oncogene family | Affinity Capture-Western | BioGRID | 15854902 |
| RHEB | MGC111559 | RHEB2 | Ras homolog enriched in brain | FRAP1 (mTOR) interacts with Rheb. | BIND | 15854902 |
| RHEB | MGC111559 | RHEB2 | Ras homolog enriched in brain | Affinity Capture-Western Reconstituted Complex | BioGRID | 15772076 |15854902 |15878852 |16915281 |
| RICTOR | DKFZp686B11164 | KIAA1999 | MGC39830 | mAVO3 | rapamycin-insensitive companion of mTOR | - | HPRD,BioGRID | 15268862 |15467718 |
| RICTOR | DKFZp686B11164 | KIAA1999 | MGC39830 | mAVO3 | rapamycin-insensitive companion of mTOR | mTOR phosphorylates mAVO3 within the mTORC2 complex. | BIND | 15467718 |
| RICTOR | DKFZp686B11164 | KIAA1999 | MGC39830 | mAVO3 | rapamycin-insensitive companion of mTOR | mTOR interacts with rictor. | BIND | 15718470 |
| RPS6KB1 | PS6K | S6K | S6K1 | STK14A | p70(S6K)-alpha | p70-S6K | p70-alpha | ribosomal protein S6 kinase, 70kDa, polypeptide 1 | Affinity Capture-Western Biochemical Activity Reconstituted Complex | BioGRID | 9465032 |10567431 |10971657 |11914378 |12150925 |12150926 |12604610 |14679009 |15809305 |15854902 |15896331 |15899889 |15905173 |16183647 |16922504 |
| STAT1 | DKFZp686B04100 | ISGF-3 | STAT91 | signal transducer and activator of transcription 1, 91kDa | Affinity Capture-Western | BioGRID | 12807916 |
| STAT3 | APRF | FLJ20882 | HIES | MGC16063 | signal transducer and activator of transcription 3 (acute-phase response factor) | Affinity Capture-Western Reconstituted Complex | BioGRID | 10660304 |15522880 |
| TERT | EST2 | TCS1 | TP2 | TRT | hEST2 | telomerase reverse transcriptase | Affinity Capture-Western | BioGRID | 15843522 |
| UBQLN1 | DA41 | DSK2 | FLJ90054 | PLIC-1 | XDRP1 | ubiquilin 1 | - | HPRD,BioGRID | 11853878 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ERBB SIGNALING PATHWAY | 87 | 71 | All SZGR 2.0 genes in this pathway |
| KEGG MTOR SIGNALING PATHWAY | 52 | 40 | All SZGR 2.0 genes in this pathway |
| KEGG INSULIN SIGNALING PATHWAY | 137 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG ADIPOCYTOKINE SIGNALING PATHWAY | 67 | 57 | All SZGR 2.0 genes in this pathway |
| KEGG TYPE II DIABETES MELLITUS | 47 | 41 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG GLIOMA | 65 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG PROSTATE CANCER | 89 | 75 | All SZGR 2.0 genes in this pathway |
| KEGG ACUTE MYELOID LEUKEMIA | 60 | 47 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BCELLSURVIVAL PATHWAY | 16 | 10 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CTCF PATHWAY | 23 | 18 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MTOR PATHWAY | 23 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA EIF4 PATHWAY | 24 | 19 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IGF1MTOR PATHWAY | 20 | 14 | All SZGR 2.0 genes in this pathway |
| SIG PIP3 SIGNALING IN CARDIAC MYOCTES | 67 | 54 | All SZGR 2.0 genes in this pathway |
| PID IL4 2PATHWAY | 65 | 43 | All SZGR 2.0 genes in this pathway |
| PID MET PATHWAY | 80 | 60 | All SZGR 2.0 genes in this pathway |
| PID IL12 2PATHWAY | 63 | 54 | All SZGR 2.0 genes in this pathway |
| PID CDC42 PATHWAY | 70 | 51 | All SZGR 2.0 genes in this pathway |
| PID LKB1 PATHWAY | 47 | 37 | All SZGR 2.0 genes in this pathway |
| PID TELOMERASE PATHWAY | 68 | 48 | All SZGR 2.0 genes in this pathway |
| PID MTOR 4PATHWAY | 69 | 55 | All SZGR 2.0 genes in this pathway |
| PID CXCR4 PATHWAY | 102 | 78 | All SZGR 2.0 genes in this pathway |
| PID IL2 PI3K PATHWAY | 34 | 27 | All SZGR 2.0 genes in this pathway |
| PID IFNG PATHWAY | 40 | 34 | All SZGR 2.0 genes in this pathway |
| PID ERBB1 DOWNSTREAM PATHWAY | 105 | 78 | All SZGR 2.0 genes in this pathway |
| PID ERBB2 ERBB3 PATHWAY | 44 | 35 | All SZGR 2.0 genes in this pathway |
| PID CXCR3 PATHWAY | 43 | 34 | All SZGR 2.0 genes in this pathway |
| PID PI3KCI AKT PATHWAY | 35 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY SCF KIT | 78 | 59 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB4 | 90 | 67 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB2 | 101 | 78 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY EGFR IN CANCER | 109 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME PI3K EVENTS IN ERBB4 SIGNALING | 38 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME PI3K EVENTS IN ERBB2 SIGNALING | 44 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | 97 | 66 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY THE B CELL RECEPTOR BCR | 126 | 90 | All SZGR 2.0 genes in this pathway |
| REACTOME INSULIN RECEPTOR SIGNALLING CASCADE | 87 | 64 | All SZGR 2.0 genes in this pathway |
| REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | 137 | 105 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR IN DISEASE | 127 | 88 | All SZGR 2.0 genes in this pathway |
| REACTOME PI3K AKT ACTIVATION | 38 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME GAB1 SIGNALOSOME | 38 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY PDGF | 122 | 93 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | 95 | 76 | All SZGR 2.0 genes in this pathway |
| REACTOME CD28 CO STIMULATION | 32 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME COSTIMULATION BY THE CD28 FAMILY | 63 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | 22 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME PI 3K CASCADE | 56 | 39 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNALING OF ACTIVATED FGFR | 100 | 74 | All SZGR 2.0 genes in this pathway |
| REACTOME PKB MEDIATED EVENTS | 29 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY INSULIN RECEPTOR | 108 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME MTORC1 MEDIATED SIGNALLING | 11 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME PIP3 ACTIVATES AKT SIGNALING | 29 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR | 112 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME PI3K CASCADE | 71 | 51 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED DN | 335 | 193 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL UP | 233 | 161 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS DN | 357 | 212 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPAIR GENES | 230 | 137 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| ASTIER INTEGRIN SIGNALING | 59 | 44 | All SZGR 2.0 genes in this pathway |
| LEE AGING NEOCORTEX DN | 80 | 49 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN DN | 153 | 120 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS DN | 371 | 218 | All SZGR 2.0 genes in this pathway |
| SU TESTIS | 76 | 53 | All SZGR 2.0 genes in this pathway |
| LEE AGING CEREBELLUM DN | 86 | 66 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS UP | 198 | 132 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN DN | 353 | 226 | All SZGR 2.0 genes in this pathway |
| DAIRKEE CANCER PRONE RESPONSE BPA | 51 | 35 | All SZGR 2.0 genes in this pathway |
| MOOTHA HUMAN MITODB 6 2002 | 429 | 260 | All SZGR 2.0 genes in this pathway |
| MOOTHA PGC | 420 | 269 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C CLUSTER UP | 38 | 26 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C UP | 47 | 29 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP C | 92 | 60 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 36HR | 29 | 23 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |