Gene Page: FRZB
Summary ?
| GeneID | 2487 |
| Symbol | FRZB |
| Synonyms | FRE|FRITZ|FRP-3|FRZB-1|FRZB-PEN|FRZB1|FZRB|OS1|SFRP3|SRFP3|hFIZ |
| Description | frizzled-related protein |
| Reference | MIM:605083|HGNC:HGNC:3959|Ensembl:ENSG00000162998|HPRD:07282|Vega:OTTHUMG00000132597 |
| Gene type | protein-coding |
| Map location | 2q32.1 |
| Pascal p-value | 0.071 |
| Sherlock p-value | 0.541 |
| Fetal beta | -0.587 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| Expression | Meta-analysis of gene expression | P value: 1.382 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs11706314 | chr3 | 45309716 | FRZB | 2487 | 0.03 | trans | ||
| rs11706394 | chr3 | 45309907 | FRZB | 2487 | 0.06 | trans | ||
| rs2976809 | chr3 | 125451738 | FRZB | 2487 | 0.05 | trans | ||
| rs6449169 | 0 | FRZB | 2487 | 0.19 | trans | |||
| rs13136254 | chr4 | 184824883 | FRZB | 2487 | 0.15 | trans | ||
| rs9313797 | chr5 | 158465457 | FRZB | 2487 | 0.18 | trans | ||
| rs17056474 | chr5 | 158468105 | FRZB | 2487 | 0.18 | trans | ||
| rs6455226 | chr6 | 67930295 | FRZB | 2487 | 0.12 | trans | ||
| rs2199402 | chr8 | 9201002 | FRZB | 2487 | 0.04 | trans | ||
| rs7841407 | chr8 | 9243427 | FRZB | 2487 | 0.01 | trans | ||
| rs17134872 | chr11 | 76139845 | FRZB | 2487 | 0.15 | trans | ||
| rs11236774 | chr11 | 76234952 | FRZB | 2487 | 0.15 | trans | ||
| rs4942043 | chr13 | 41978218 | FRZB | 2487 | 0.14 | trans | ||
| rs4402447 | chr13 | 41984741 | FRZB | 2487 | 0.19 | trans | ||
| rs9529974 | chr13 | 72821935 | FRZB | 2487 | 0.13 | trans | ||
| rs17720221 | chr13 | 75182377 | FRZB | 2487 | 0.06 | trans | ||
| rs6095741 | chr20 | 48666589 | FRZB | 2487 | 0.11 | trans | ||
| rs5991662 | chrX | 43335796 | FRZB | 2487 | 0.03 | trans |
Section II. Transcriptome annotation
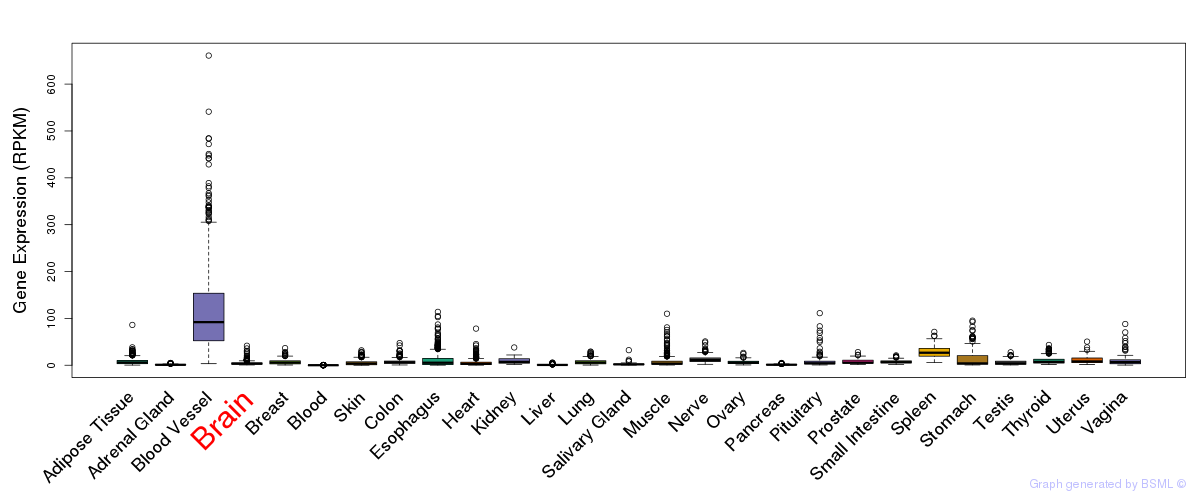
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0017147 | Wnt-protein binding | ISS | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001501 | skeletal system development | TAS | 8824257 | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| GO:0030178 | negative regulation of Wnt receptor signaling pathway | ISS | - | |
| GO:0030154 | cell differentiation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | TAS | 9096311 | |
| GO:0016020 | membrane | TAS | 8824257 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| WNT SIGNALING | 89 | 71 | All SZGR 2.0 genes in this pathway |
| WINTER HYPOXIA DN | 52 | 30 | All SZGR 2.0 genes in this pathway |
| BORCZUK MALIGNANT MESOTHELIOMA DN | 104 | 59 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| LIEN BREAST CARCINOMA METAPLASTIC VS DUCTAL DN | 114 | 58 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 2 DN | 51 | 42 | All SZGR 2.0 genes in this pathway |
| HATADA METHYLATED IN LUNG CANCER UP | 390 | 236 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 UP | 209 | 139 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA HIF1A UP | 77 | 49 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 AND HIF1A DN | 103 | 71 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY UP | 236 | 139 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS UP | 344 | 180 | All SZGR 2.0 genes in this pathway |
| RICKMAN HEAD AND NECK CANCER A | 100 | 63 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| SAKAI CHRONIC HEPATITIS VS LIVER CANCER DN | 36 | 24 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA HP UP | 49 | 26 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE UP | 244 | 165 | All SZGR 2.0 genes in this pathway |
| MACLACHLAN BRCA1 TARGETS DN | 16 | 12 | All SZGR 2.0 genes in this pathway |
| SANSOM WNT PATHWAY REQUIRE MYC | 58 | 43 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY GAMMA RADIATION | 81 | 59 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE UP | 226 | 164 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| YAUCH HEDGEHOG SIGNALING PARACRINE UP | 149 | 85 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS UP | 165 | 118 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS DN | 138 | 92 | All SZGR 2.0 genes in this pathway |
| DAVIES MULTIPLE MYELOMA VS MGUS UP | 13 | 10 | All SZGR 2.0 genes in this pathway |
| FINAK BREAST CANCER SDPP SIGNATURE | 26 | 11 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| CROONQUIST IL6 DEPRIVATION DN | 98 | 69 | All SZGR 2.0 genes in this pathway |
| ZHAN V1 LATE DIFFERENTIATION GENES DN | 15 | 13 | All SZGR 2.0 genes in this pathway |
| POOLA INVASIVE BREAST CANCER DN | 134 | 83 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER LATE RECURRENCE UP | 62 | 42 | All SZGR 2.0 genes in this pathway |
| FONTAINE THYROID TUMOR UNCERTAIN MALIGNANCY DN | 26 | 14 | All SZGR 2.0 genes in this pathway |
| FONTAINE PAPILLARY THYROID CARCINOMA DN | 80 | 53 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE G2 M | 216 | 124 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT | 567 | 365 | All SZGR 2.0 genes in this pathway |
| WANG MLL TARGETS | 289 | 188 | All SZGR 2.0 genes in this pathway |
| ROLEF GLIS3 TARGETS | 37 | 29 | All SZGR 2.0 genes in this pathway |
| ALFANO MYC TARGETS | 239 | 156 | All SZGR 2.0 genes in this pathway |
| NABA SECRETED FACTORS | 344 | 197 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-338 | 55 | 61 | 1A | hsa-miR-338brain | UCCAGCAUCAGUGAUUUUGUUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.