Gene Page: GABRB3
Summary ?
| GeneID | 2562 |
| Symbol | GABRB3 |
| Synonyms | ECA5 |
| Description | gamma-aminobutyric acid type A receptor beta3 subunit |
| Reference | MIM:137192|HGNC:HGNC:4083|Ensembl:ENSG00000166206|HPRD:08844|Vega:OTTHUMG00000129231 |
| Gene type | protein-coding |
| Map location | 15q12 |
| Pascal p-value | 5.533E-4 |
| Sherlock p-value | 0.617 |
| Fetal beta | -0.705 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
| Support | LIGAND GATED ION SIGNALING Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CNV:YES | Copy number variation studies | Manual curation | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 4 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg13904560 | 15 | 26870826 | GABRB3 | 1.18E-6 | 0.583 | 0.007 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1155107 | chr3 | 26843376 | GABRB3 | 2562 | 0.1 | trans | ||
| rs17029291 | chr3 | 32402138 | GABRB3 | 2562 | 0.02 | trans | ||
| rs750544 | chr5 | 51749054 | GABRB3 | 2562 | 0.14 | trans | ||
| rs240410 | chr6 | 75940738 | GABRB3 | 2562 | 0.18 | trans | ||
| rs418485 | chr6 | 75966268 | GABRB3 | 2562 | 0.18 | trans | ||
| rs240388 | chr6 | 75972698 | GABRB3 | 2562 | 0.18 | trans | ||
| rs1324669 | chr13 | 107872446 | GABRB3 | 2562 | 0.01 | trans | ||
| rs17145698 | chrX | 40218345 | GABRB3 | 2562 | 0.03 | trans |
Section II. Transcriptome annotation
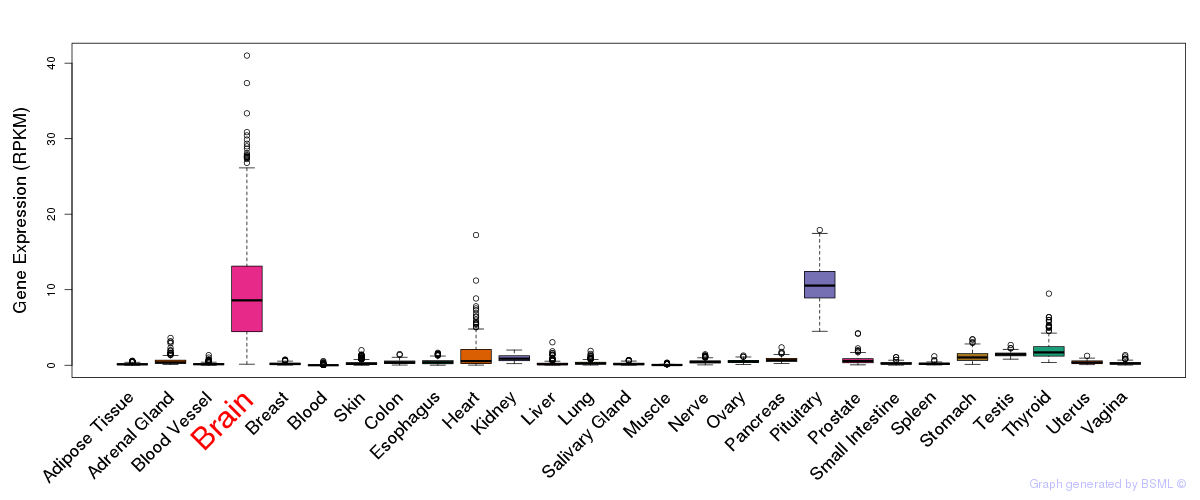
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0030594 | neurotransmitter receptor activity | IEA | Neurotransmitter (GO term level: 5) | - |
| GO:0004890 | GABA-A receptor activity | IEA | GABA (GO term level: 7) | - |
| GO:0004890 | GABA-A receptor activity | TAS | GABA (GO term level: 7) | 1664410 |
| GO:0005254 | chloride channel activity | IEA | - | |
| GO:0005216 | ion channel activity | IEA | - | |
| GO:0005230 | extracellular ligand-gated ion channel activity | IEA | - | |
| GO:0031404 | chloride ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007165 | signal transduction | TAS | 1664410 | |
| GO:0006811 | ion transport | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0045211 | postsynaptic membrane | IEA | Synap, Neurotransmitter (GO term level: 5) | - |
| GO:0045202 | synapse | IEA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 1664410 | |
| GO:0030054 | cell junction | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | 186 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| REACTOME NEUROTRANSMITTER RECEPTOR BINDING AND DOWNSTREAM TRANSMISSION IN THE POSTSYNAPTIC CELL | 137 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMEMBRANE TRANSPORT OF SMALL MOLECULES | 413 | 270 | All SZGR 2.0 genes in this pathway |
| REACTOME GABA A RECEPTOR ACTIVATION | 12 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME GABA RECEPTOR ACTIVATION | 52 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME ION CHANNEL TRANSPORT | 55 | 42 | All SZGR 2.0 genes in this pathway |
| REACTOME LIGAND GATED ION CHANNEL TRANSPORT | 21 | 21 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY DN | 367 | 220 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS DN | 536 | 332 | All SZGR 2.0 genes in this pathway |
| ODONNELL METASTASIS UP | 82 | 58 | All SZGR 2.0 genes in this pathway |
| HUMMERICH SKIN CANCER PROGRESSION UP | 88 | 58 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES 1 | 379 | 235 | All SZGR 2.0 genes in this pathway |
| POMEROY MEDULLOBLASTOMA DESMOPLASIC VS CLASSIC UP | 62 | 38 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN DN | 153 | 120 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE UP | 249 | 170 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT DN | 165 | 106 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS UP | 170 | 107 | All SZGR 2.0 genes in this pathway |
| EHLERS ANEUPLOIDY DN | 12 | 8 | All SZGR 2.0 genes in this pathway |
| CONRAD STEM CELL | 39 | 27 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR DN | 36 | 19 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 AND SATB1 DN | 180 | 116 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-128 | 2301 | 2307 | 1A | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-194 | 2904 | 2910 | m8 | hsa-miR-194 | UGUAACAGCAACUCCAUGUGGA |
| miR-203.1 | 2670 | 2676 | 1A | hsa-miR-203 | UGAAAUGUUUAGGACCACUAG |
| miR-204/211 | 329 | 335 | m8 | hsa-miR-204brain | UUCCCUUUGUCAUCCUAUGCCU |
| hsa-miR-211 | UUCCCUUUGUCAUCCUUCGCCU | ||||
| miR-218 | 4121 | 4127 | m8 | hsa-miR-218brain | UUGUGCUUGAUCUAACCAUGU |
| hsa-miR-218brain | UUGUGCUUGAUCUAACCAUGU | ||||
| miR-23 | 136 | 142 | 1A | hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC |
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| miR-26 | 1798 | 1804 | m8 | hsa-miR-26abrain | UUCAAGUAAUCCAGGAUAGGC |
| hsa-miR-26bSZ | UUCAAGUAAUUCAGGAUAGGUU | ||||
| miR-27 | 2301 | 2307 | m8 | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-369-3p | 4144 | 4150 | m8 | hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU |
| miR-383 | 1362 | 1368 | m8 | hsa-miR-383brain | AGAUCAGAAGGUGAUUGUGGCU |
| miR-421 | 1312 | 1318 | 1A | hsa-miR-421 | GGCCUCAUUAAAUGUUUGUUG |
| miR-431 | 1411 | 1417 | 1A | hsa-miR-431 | UGUCUUGCAGGCCGUCAUGCA |
| miR-448 | 2479 | 2485 | m8 | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
| miR-455 | 3486 | 3492 | m8 | hsa-miR-455 | UAUGUGCCUUUGGACUACAUCG |
| miR-485-5p | 1371 | 1378 | 1A,m8 | hsa-miR-485-5p | AGAGGCUGGCCGUGAUGAAUUC |
| miR-486 | 4021 | 4028 | 1A,m8 | hsa-miR-486 | UCCUGUACUGAGCUGCCCCGAG |
| miR-488 | 1987 | 1993 | 1A | hsa-miR-488 | CCCAGAUAAUGGCACUCUCAA |
| miR-500 | 1418 | 1424 | m8 | hsa-miR-500 | AUGCACCUGGGCAAGGAUUCUG |
| miR-539 | 144 | 150 | 1A | hsa-miR-539 | GGAGAAAUUAUCCUUGGUGUGU |
| miR-542-3p | 1566 | 1572 | 1A | hsa-miR-542-3p | UGUGACAGAUUGAUAACUGAAA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.