Gene Page: TINF2
Summary ?
| GeneID | 26277 |
| Symbol | TINF2 |
| Synonyms | DKCA3|TIN2 |
| Description | TERF1 (TRF1)-interacting nuclear factor 2 |
| Reference | MIM:604319|HGNC:HGNC:11824|Ensembl:ENSG00000092330|HPRD:16056|Vega:OTTHUMG00000171849 |
| Gene type | protein-coding |
| Map location | 14q12 |
| Pascal p-value | 0.164 |
| Sherlock p-value | 0.334 |
| TADA p-value | 0.009 |
| Fetal beta | 0.327 |
| eGene | Myers' cis & trans |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.047 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| TINF2 | chr14 | 24709311 | A | C | NM_001099274 NM_012461 | p.394L>V . | missense 3-UTR | Schizophrenia | DNM:Fromer_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1111763 | chr14 | 25591536 | TINF2 | 26277 | 0.09 | cis |
Section II. Transcriptome annotation
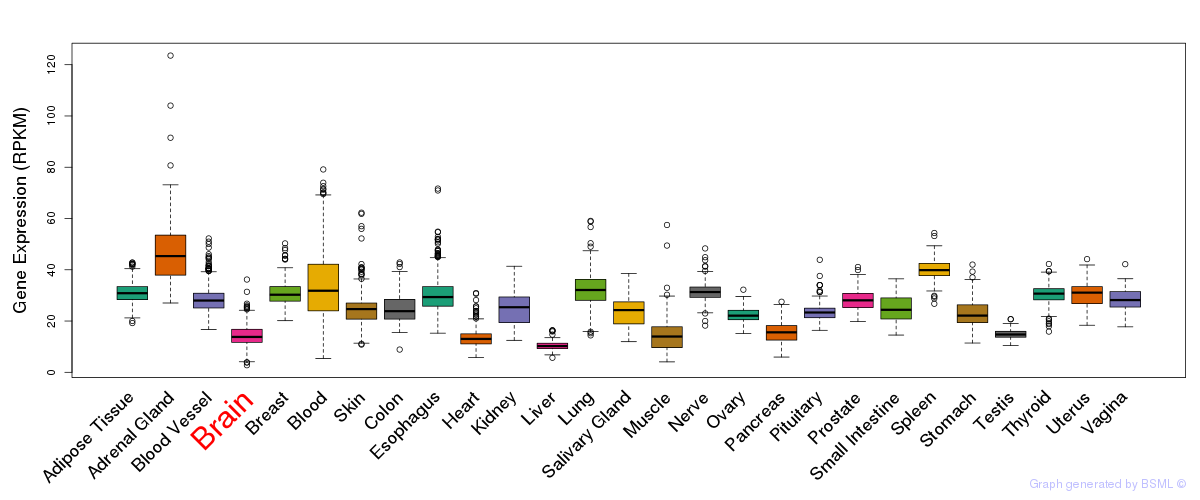
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003677 | DNA binding | TAS | 10581025 | |
| GO:0005515 | protein binding | IPI | 12768206 | |
| GO:0042162 | telomeric DNA binding | IDA | 12768206 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007004 | telomere maintenance via telomerase | TAS | 10581025 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000781 | chromosome, telomeric region | TAS | 10581025 | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005654 | nucleoplasm | EXP | 16166375 | |
| GO:0005694 | chromosome | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID TELOMERASE PATHWAY | 68 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME MEIOSIS | 116 | 81 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME CHROMOSOME MAINTENANCE | 122 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME MEIOTIC SYNAPSIS | 73 | 57 | All SZGR 2.0 genes in this pathway |
| REACTOME PACKAGING OF TELOMERE ENDS | 48 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME TELOMERE MAINTENANCE | 75 | 57 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| JAZAG TGFB1 SIGNALING UP | 108 | 69 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY DN | 382 | 224 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPAIR GENES | 230 | 137 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL LONG TERM | 302 | 191 | All SZGR 2.0 genes in this pathway |
| HU GENOTOXIN ACTION DIRECT VS INDIRECT 24HR | 55 | 38 | All SZGR 2.0 genes in this pathway |
| BILD MYC ONCOGENIC SIGNATURE | 206 | 117 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| CONRAD STEM CELL | 39 | 27 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE EARLY LATE | 317 | 190 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |