Gene Page: GNB3
Summary ?
| GeneID | 2784 |
| Symbol | GNB3 |
| Synonyms | - |
| Description | G protein subunit beta 3 |
| Reference | MIM:139130|HGNC:HGNC:4400|Ensembl:ENSG00000111664|HPRD:00743|Vega:OTTHUMG00000168517 |
| Gene type | protein-coding |
| Map location | 12p13 |
| Pascal p-value | 0.205 |
| Sherlock p-value | 0.888 |
| eGene | Caudate basal ganglia Cerebellar Hemisphere Cerebellum Cortex Hippocampus Hypothalamus Nucleus accumbens basal ganglia Putamen basal ganglia Myers' cis & trans Meta |
| Support | CANABINOID DOPAMINE G-PROTEIN RELAY METABOTROPIC GLUTAMATE RECEPTOR SEROTONIN |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 1 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs3917788 | chr1 | 169568718 | GNB3 | 2784 | 0.18 | trans | ||
| rs6666913 | chr1 | 237092144 | GNB3 | 2784 | 2.115E-4 | trans | ||
| rs16865199 | chr2 | 6621910 | GNB3 | 2784 | 0.18 | trans | ||
| rs9870631 | chr3 | 29583547 | GNB3 | 2784 | 0.12 | trans | ||
| rs2324466 | chr3 | 76100753 | GNB3 | 2784 | 0 | trans | ||
| rs17013713 | chr3 | 76105150 | GNB3 | 2784 | 3.197E-4 | trans | ||
| rs17013722 | chr3 | 76107656 | GNB3 | 2784 | 0 | trans | ||
| rs17013724 | chr3 | 76107684 | GNB3 | 2784 | 0 | trans | ||
| rs7644174 | chr3 | 112722325 | GNB3 | 2784 | 0.05 | trans | ||
| rs9882442 | chr3 | 173644484 | GNB3 | 2784 | 0.19 | trans | ||
| rs11937178 | chr4 | 148847598 | GNB3 | 2784 | 0 | trans | ||
| rs11934409 | chr4 | 148848884 | GNB3 | 2784 | 0 | trans | ||
| rs9986296 | chr5 | 3199318 | GNB3 | 2784 | 0 | trans | ||
| rs17463444 | chr5 | 95174483 | GNB3 | 2784 | 0.11 | trans | ||
| rs1030668 | chr5 | 166876426 | GNB3 | 2784 | 0.06 | trans | ||
| rs992435 | chr7 | 97050802 | GNB3 | 2784 | 0.18 | trans | ||
| rs8190856 | chr8 | 18079123 | GNB3 | 2784 | 0.1 | trans | ||
| rs16920368 | chr8 | 55392852 | GNB3 | 2784 | 0.07 | trans | ||
| rs348150 | chr8 | 73371619 | GNB3 | 2784 | 0.04 | trans | ||
| rs16938220 | chr8 | 73377940 | GNB3 | 2784 | 0.04 | trans | ||
| rs16908622 | chr8 | 139231604 | GNB3 | 2784 | 0.06 | trans | ||
| rs4878005 | chr9 | 89316567 | GNB3 | 2784 | 0.15 | trans | ||
| rs12685165 | chr9 | 89340049 | GNB3 | 2784 | 0.1 | trans | ||
| rs12686427 | chr9 | 89340546 | GNB3 | 2784 | 0.1 | trans | ||
| rs10119434 | chr9 | 110944789 | GNB3 | 2784 | 0.11 | trans | ||
| rs10982951 | chr9 | 118609806 | GNB3 | 2784 | 0.02 | trans | ||
| rs11193467 | chr10 | 109217296 | GNB3 | 2784 | 0.14 | trans | ||
| rs1002405 | chr11 | 85572958 | GNB3 | 2784 | 0.05 | trans | ||
| rs2069410 | chr12 | 56364644 | GNB3 | 2784 | 0.04 | trans | ||
| rs17834315 | chr13 | 37991091 | GNB3 | 2784 | 0.09 | trans | ||
| rs11841288 | chr13 | 60215833 | GNB3 | 2784 | 0.16 | trans | ||
| rs10498433 | chr14 | 52289530 | GNB3 | 2784 | 0 | trans | ||
| rs1743505 | chr14 | 96143407 | GNB3 | 2784 | 0.06 | trans | ||
| rs16946913 | chr15 | 63653889 | GNB3 | 2784 | 0.1 | trans | ||
| snp_a-4250222 | 0 | GNB3 | 2784 | 0.04 | trans | |||
| rs6499075 | chr16 | 66387395 | GNB3 | 2784 | 0.03 | trans | ||
| rs2826625 | chr21 | 22341050 | GNB3 | 2784 | 0.12 | trans | ||
| rs2826648 | chr21 | 22362498 | GNB3 | 2784 | 0.12 | trans | ||
| rs9980476 | chr21 | 22365306 | GNB3 | 2784 | 0.12 | trans | ||
| rs1041786 | chr21 | 22617710 | GNB3 | 2784 | 0.02 | trans | ||
| rs7055735 | chrX | 85687566 | GNB3 | 2784 | 0.06 | trans | ||
| rs1129649 | 12 | 6948468 | GNB3 | ENSG00000111664.6 | 2.596E-7 | 0.01 | -907 | gtex_brain_putamen_basal |
| rs5440 | 12 | 6948899 | GNB3 | ENSG00000111664.6 | 2.215E-6 | 0.01 | -476 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
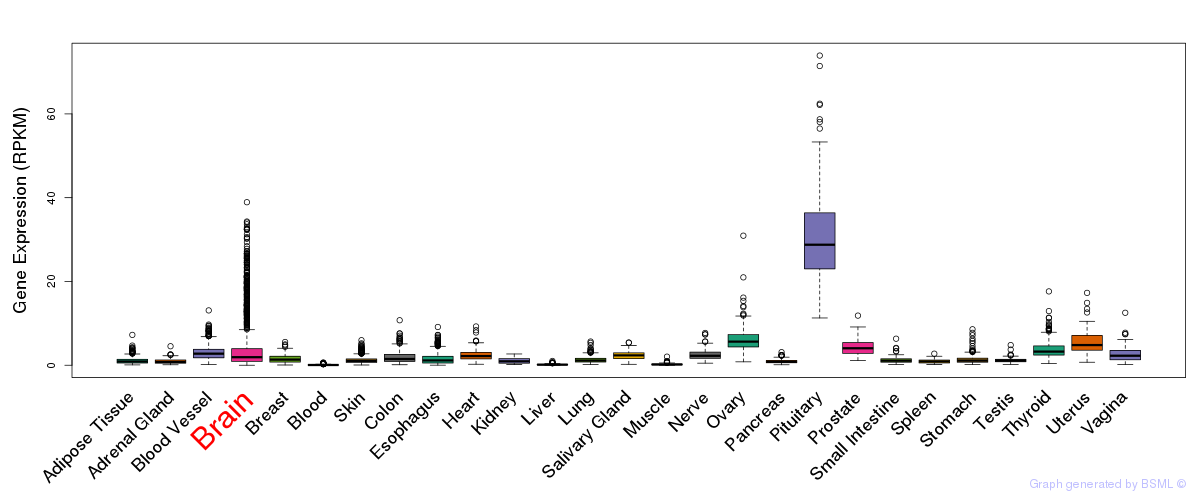
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AL117209.1 | 0.91 | 0.90 |
| AP3D1 | 0.91 | 0.90 |
| EIF4G1 | 0.91 | 0.89 |
| GRIPAP1 | 0.90 | 0.87 |
| MPRIP | 0.89 | 0.90 |
| LARP1 | 0.89 | 0.89 |
| ANKRD11 | 0.89 | 0.90 |
| GOLGA3 | 0.89 | 0.92 |
| VPS53 | 0.89 | 0.88 |
| HERC2 | 0.89 | 0.89 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.75 | -0.71 |
| C1orf54 | -0.73 | -0.73 |
| AF347015.31 | -0.71 | -0.66 |
| GNG11 | -0.69 | -0.67 |
| VAMP5 | -0.68 | -0.62 |
| HIGD1B | -0.68 | -0.63 |
| MT-CO2 | -0.67 | -0.62 |
| ENHO | -0.66 | -0.60 |
| SAT1 | -0.65 | -0.62 |
| S100A13 | -0.65 | -0.63 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003993 | acid phosphatase activity | IEA | - | |
| GO:0004871 | signal transducer activity | IEA | - | |
| GO:0003924 | GTPase activity | TAS | 9425898 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007186 | G-protein coupled receptor protein signaling pathway | TAS | 2107550 | |
| GO:0008217 | regulation of blood pressure | TAS | 9425898 | |
| GO:0007165 | signal transduction | IEA | - | |
| GO:0009755 | hormone-mediated signaling | EXP | 12626323 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IDA | 18029348 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CHEMOKINE SIGNALING PATHWAY | 190 | 128 | All SZGR 2.0 genes in this pathway |
| KEGG TASTE TRANSDUCTION | 52 | 28 | All SZGR 2.0 genes in this pathway |
| PID CONE PATHWAY | 23 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME GASTRIN CREB SIGNALLING PATHWAY VIA PKC AND MAPK | 205 | 136 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | 186 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRATION OF ENERGY METABOLISM | 120 | 84 | All SZGR 2.0 genes in this pathway |
| REACTOME OPIOID SIGNALLING | 78 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME NEUROTRANSMITTER RECEPTOR BINDING AND DOWNSTREAM TRANSMISSION IN THE POSTSYNAPTIC CELL | 137 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME G PROTEIN ACTIVATION | 27 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMEMBRANE TRANSPORT OF SMALL MOLECULES | 413 | 270 | All SZGR 2.0 genes in this pathway |
| REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | 34 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | 43 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA Q SIGNALLING EVENTS | 184 | 116 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF INSULIN SECRETION | 93 | 65 | All SZGR 2.0 genes in this pathway |
| REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | 25 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | 88 | 58 | All SZGR 2.0 genes in this pathway |
| REACTOME GLUCAGON TYPE LIGAND RECEPTORS | 33 | 22 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA1213 SIGNALLING EVENTS | 74 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME ADP SIGNALLING THROUGH P2RY1 | 25 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | 20 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA I SIGNALLING EVENTS | 195 | 114 | All SZGR 2.0 genes in this pathway |
| REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | 25 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA S SIGNALLING EVENTS | 121 | 82 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA Z SIGNALLING EVENTS | 44 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME G PROTEIN BETA GAMMA SIGNALLING | 28 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNAL AMPLIFICATION | 31 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | 23 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME ADP SIGNALLING THROUGH P2RY12 | 21 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | 31 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR LIGAND BINDING | 408 | 246 | All SZGR 2.0 genes in this pathway |
| REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | 32 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET HOMEOSTASIS | 78 | 49 | All SZGR 2.0 genes in this pathway |
| REACTOME AQUAPORIN MEDIATED TRANSPORT | 51 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | 19 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME INCRETIN SYNTHESIS SECRETION AND INACTIVATION | 22 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | 19 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | 44 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | 25 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME GABA B RECEPTOR ACTIVATION | 38 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME GABA RECEPTOR ACTIVATION | 52 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME POTASSIUM CHANNELS | 98 | 68 | All SZGR 2.0 genes in this pathway |
| REACTOME INWARDLY RECTIFYING K CHANNELS | 31 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | 208 | 138 | All SZGR 2.0 genes in this pathway |
| KORKOLA SEMINOMA UP | 44 | 27 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 1 DN | 378 | 231 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 UP | 329 | 196 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| PENG GLUTAMINE DEPRIVATION UP | 38 | 25 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| POMEROY MEDULLOBLASTOMA DESMOPLASIC VS CLASSIC UP | 62 | 38 | All SZGR 2.0 genes in this pathway |
| PENG RAPAMYCIN RESPONSE UP | 203 | 130 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE UP | 244 | 165 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 8HR UP | 105 | 73 | All SZGR 2.0 genes in this pathway |
| LABBE TARGETS OF TGFB1 AND WNT3A DN | 108 | 68 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| ZHAN VARIABLE EARLY DIFFERENTIATION GENES UP | 15 | 9 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN | 841 | 431 | All SZGR 2.0 genes in this pathway |
| ZWANG EGF INTERVAL UP | 105 | 46 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-221/222 | 481 | 487 | 1A | hsa-miR-221brain | AGCUACAUUGUCUGCUGGGUUUC |
| hsa-miR-222brain | AGCUACAUCUGGCUACUGGGUCUC | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.