Gene Page: GRK6
Summary ?
| GeneID | 2870 |
| Symbol | GRK6 |
| Synonyms | GPRK6 |
| Description | G protein-coupled receptor kinase 6 |
| Reference | MIM:600869|HGNC:HGNC:4545|Ensembl:ENSG00000198055|HPRD:02925|Vega:OTTHUMG00000163401 |
| Gene type | protein-coding |
| Map location | 5q35 |
| Pascal p-value | 0.272 |
| Fetal beta | -0.012 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.0276 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
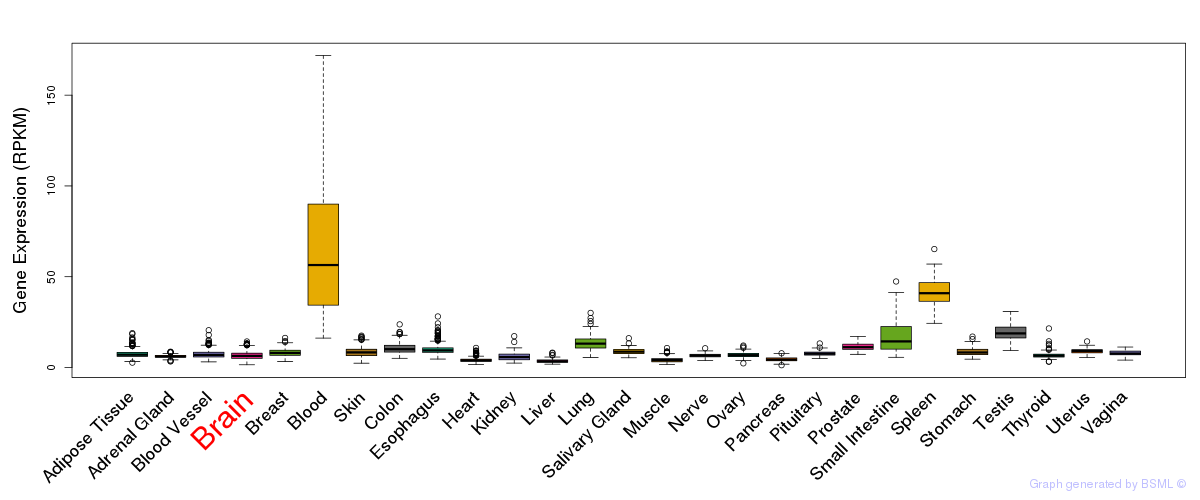
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ADAR | 0.88 | 0.90 |
| LARP1 | 0.87 | 0.90 |
| EIF2AK4 | 0.86 | 0.88 |
| KIAA0100 | 0.86 | 0.89 |
| LARP4B | 0.86 | 0.89 |
| MTOR | 0.85 | 0.88 |
| PRDM4 | 0.85 | 0.88 |
| HLCS | 0.85 | 0.86 |
| CLCN6 | 0.85 | 0.87 |
| KPNA6 | 0.84 | 0.88 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.68 | -0.68 |
| AF347015.31 | -0.66 | -0.65 |
| MT-CO2 | -0.64 | -0.64 |
| HIGD1B | -0.63 | -0.63 |
| AF347015.8 | -0.63 | -0.63 |
| AF347015.2 | -0.62 | -0.58 |
| C1orf54 | -0.61 | -0.68 |
| PLA2G5 | -0.60 | -0.61 |
| FXYD1 | -0.60 | -0.58 |
| MT-CYB | -0.60 | -0.58 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0004871 | signal transducer activity | IEA | - | |
| GO:0004703 | G-protein coupled receptor kinase activity | IEA | - | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| GO:0007165 | signal transduction | IEA | - | |
| GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway | TAS | 8077221 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016020 | membrane | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ADRB2 | ADRB2R | ADRBR | B2AR | BAR | BETA2AR | adrenergic, beta-2-, receptor, surface | - | HPRD,BioGRID | 9819198 |
| CHRM3 | HM3 | cholinergic receptor, muscarinic 3 | - | HPRD | 11856737 |
| EDNRB | ABCDS | ETB | ETBR | ETRB | HSCR | HSCR2 | endothelin receptor type B | - | HPRD | 9211925 |
| FSHR | FSHRO | LGR1 | MGC141667 | MGC141668 | ODG1 | follicle stimulating hormone receptor | - | HPRD,BioGRID | 10379886 |
| GIT1 | - | G protein-coupled receptor kinase interacting ArfGAP 1 | - | HPRD | 9826657 |
| PRSS23 | MGC5107 | SIG13 | SPUVE | ZSIG13 | protease, serine, 23 | Two-hybrid | BioGRID | 16169070 |
| RCVRN | RCV1 | recoverin | - | HPRD,BioGRID | 12507501 |
| RHO | CSNBAD1 | MGC138309 | MGC138311 | OPN2 | RP4 | rhodopsin | - | HPRD | 8366096 |
| SNCA | MGC110988 | NACP | PARK1 | PARK4 | PD1 | synuclein, alpha (non A4 component of amyloid precursor) | Biochemical Activity | BioGRID | 10852916 |
| SNCB | - | synuclein, beta | Biochemical Activity | BioGRID | 10852916 |
| SNCG | BCSG1 | SR | synuclein, gamma (breast cancer-specific protein 1) | Biochemical Activity | BioGRID | 10852916 |
| TBXA2R | TXA2-R | thromboxane A2 receptor | - | HPRD | 11504827 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CHEMOKINE SIGNALING PATHWAY | 190 | 128 | All SZGR 2.0 genes in this pathway |
| KEGG ENDOCYTOSIS | 183 | 132 | All SZGR 2.0 genes in this pathway |
| PID CXCR4 PATHWAY | 102 | 78 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 UP | 201 | 125 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 UP | 418 | 263 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| PROVENZANI METASTASIS DN | 136 | 94 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS UP | 293 | 179 | All SZGR 2.0 genes in this pathway |
| ODONNELL METASTASIS DN | 24 | 13 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| LIAO METASTASIS | 539 | 324 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| SINGH NFE2L2 TARGETS | 15 | 12 | All SZGR 2.0 genes in this pathway |
| ZHENG BOUND BY FOXP3 | 491 | 310 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER POOR SURVIVAL DN | 22 | 15 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| FIRESTEIN PROLIFERATION | 175 | 125 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE G2 M | 216 | 124 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION UP | 271 | 165 | All SZGR 2.0 genes in this pathway |
| FOSTER KDM1A TARGETS DN | 211 | 119 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-1/206 | 752 | 758 | m8 | hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA |
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| miR-137 | 792 | 798 | 1A | hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG |
| miR-181 | 225 | 231 | 1A | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-19 | 28 | 35 | 1A,m8 | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-22 | 1 | 7 | 1A | hsa-miR-22brain | AAGCUGCCAGUUGAAGAACUGU |
| miR-34/449 | 600 | 606 | m8 | hsa-miR-34abrain | UGGCAGUGUCUUAGCUGGUUGUU |
| hsa-miR-34c | AGGCAGUGUAGUUAGCUGAUUGC | ||||
| hsa-miR-449 | UGGCAGUGUAUUGUUAGCUGGU | ||||
| hsa-miR-449b | AGGCAGUGUAUUGUUAGCUGGC | ||||
| miR-409-3p | 751 | 757 | m8 | hsa-miR-409-3p | CGAAUGUUGCUCGGUGAACCCCU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.