Gene Page: HIPK2
Summary ?
| GeneID | 28996 |
| Symbol | HIPK2 |
| Synonyms | PRO0593 |
| Description | homeodomain interacting protein kinase 2 |
| Reference | MIM:606868|HGNC:HGNC:14402|Ensembl:ENSG00000064393|HPRD:06039|Vega:OTTHUMG00000157704 |
| Gene type | protein-coding |
| Map location | 7q34 |
| Pascal p-value | 0.006 |
| Sherlock p-value | 3.539E-5 |
| Fetal beta | -0.935 |
| eGene | Myers' cis & trans |
| Support | CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs483323 | chr1 | 107958416 | HIPK2 | 28996 | 0.07 | trans | ||
| rs7340968 | chr4 | 78961665 | HIPK2 | 28996 | 0.15 | trans | ||
| rs17614208 | chr4 | 165147508 | HIPK2 | 28996 | 0.19 | trans |
Section II. Transcriptome annotation
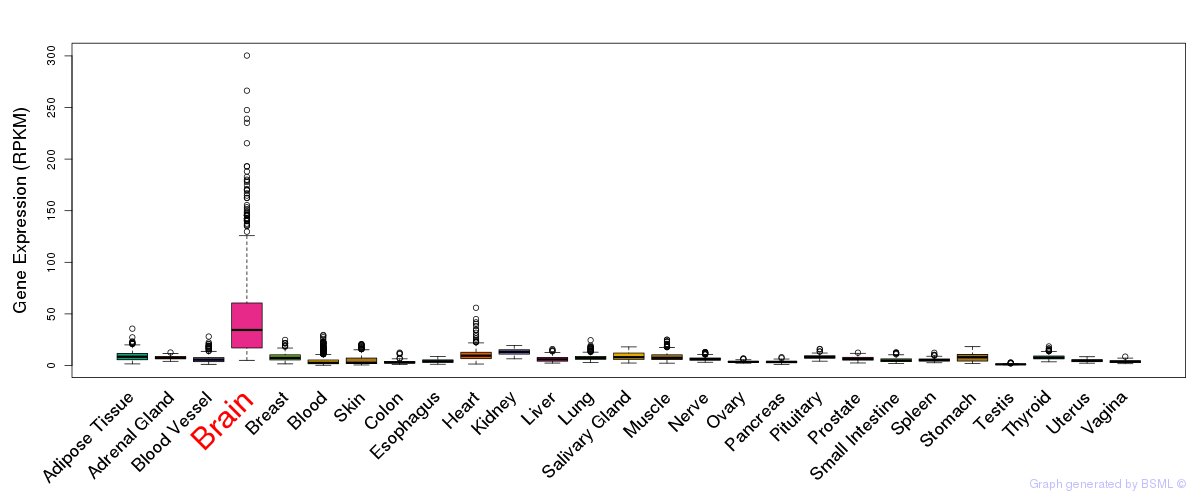
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | - | HPRD,BioGRID | 11740489 |
| DAXX | BING2 | DAP6 | EAP1 | MGC126245 | MGC126246 | death-domain associated protein | - | HPRD,BioGRID | 14678985 |
| HMGA1 | HMG-R | HMGA1A | HMGIY | MGC12816 | MGC4242 | MGC4854 | high mobility group AT-hook 1 | - | HPRD | 11593421 |
| NKX2-1 | BCH | BHC | NK-2 | NKX2.1 | NKX2A | TEBP | TITF1 | TTF-1 | TTF1 | NK2 homeobox 1 | - | HPRD,BioGRID | 11438542 |
| NKX2-5 | CHNG5 | CSX | CSX1 | NKX2.5 | NKX2E | NKX4-1 | NK2 transcription factor related, locus 5 (Drosophila) | - | HPRD | 9748262 |
| NLK | DKFZp761G1211 | FLJ21033 | nemo-like kinase | - | HPRD | 15082531 |
| PTCH1 | BCNS | FLJ26746 | FLJ42602 | HPE7 | NBCCS | PTC | PTC1 | PTCH | PTCH11 | patched homolog 1 (Drosophila) | - | HPRD | 12874272 |
| RANBP9 | RANBPM | RAN binding protein 9 | The nuclear protein kinase HIPK2 interacts with the Ran binding protein RanBPM | BIND | 12220523 |
| RANBP9 | RANBPM | RAN binding protein 9 | - | HPRD,BioGRID | 12220523 |
| SKI | SKV | v-ski sarcoma viral oncogene homolog (avian) | - | HPRD,BioGRID | 12874272 |
| SMAD1 | BSP1 | JV4-1 | JV41 | MADH1 | MADR1 | SMAD family member 1 | - | HPRD,BioGRID | 12874272 |
| SMAD2 | JV18 | JV18-1 | MADH2 | MADR2 | MGC22139 | MGC34440 | hMAD-2 | hSMAD2 | SMAD family member 2 | - | HPRD,BioGRID | 12874272 |
| SMAD3 | DKFZp586N0721 | DKFZp686J10186 | HSPC193 | HsT17436 | JV15-2 | MADH3 | MGC60396 | SMAD family member 3 | Reconstituted Complex | BioGRID | 12874272 |
| SP100 | DKFZp686E07254 | FLJ00340 | FLJ34579 | SP100 nuclear antigen | - | HPRD,BioGRID | 14647468 |
| SUMO1 | DAP-1 | GMP1 | OFC10 | PIC1 | SENP2 | SMT3 | SMT3C | SMT3H3 | SUMO-1 | UBL1 | SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae) | SUMO-1 interacts with HIPK2. | BIND | 14609633 |
| SUMO1 | DAP-1 | GMP1 | OFC10 | PIC1 | SENP2 | SMT3 | SMT3C | SMT3H3 | SUMO-1 | UBL1 | SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae) | - | HPRD,BioGRID | 14609633 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | Affinity Capture-Western Biochemical Activity Reconstituted Complex Two-hybrid | BioGRID | 11740489 |11925430 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | HIPK2 does not interact directly with tumour suppressor protein p53 but enhances p53 activity and p53 expression or stability | BIND | 11532197 |
| TP53INP1 | DKFZp434M1317 | FLJ22139 | SIP | TP53DINP1 | TP53INP1A | TP53INP1B | Teap | p53DINP1 | tumor protein p53 inducible nuclear protein 1 | - | HPRD,BioGRID | 12851404 |
| TP63 | AIS | B(p51A) | B(p51B) | EEC3 | KET | LMS | NBP | OFC8 | RHS | SHFM4 | TP53CP | TP53L | TP73L | p40 | p51 | p53CP | p63 | p73H | p73L | tumor protein p63 | - | HPRD,BioGRID | 11925430 |
| TP73 | P73 | tumor protein p73 | - | HPRD,BioGRID | 11925430 |
| TRADD | Hs.89862 | MGC11078 | TNFRSF1A-associated via death domain | - | HPRD,BioGRID | 11032752 |
| UBE2I | C358B7.1 | P18 | UBC9 | ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) | Ubc9 with HIPK2. | BIND | 14609633 |
| UBE2I | C358B7.1 | P18 | UBC9 | ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) | Two-hybrid | BioGRID | 14609633 |
| UBE2I | C358B7.1 | P18 | UBC9 | ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) | - | HPRD | 10535925 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID CMYB PATHWAY | 84 | 61 | All SZGR 2.0 genes in this pathway |
| PID P53 REGULATION PATHWAY | 59 | 50 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST LOBULAR CARCINOMA VS DUCTAL NORMAL DN | 91 | 53 | All SZGR 2.0 genes in this pathway |
| WILCOX RESPONSE TO PROGESTERONE DN | 66 | 44 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS UP | 579 | 346 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE UP | 485 | 293 | All SZGR 2.0 genes in this pathway |
| WANG LMO4 TARGETS UP | 372 | 227 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| ODONNELL TFRC TARGETS UP | 456 | 228 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA UP | 544 | 308 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LPS UP | 431 | 237 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA DN | 77 | 52 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND SERUM DEPRIVATION UP | 211 | 136 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 4 5WK DN | 196 | 131 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 6 7WK UP | 197 | 135 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 2 | 127 | 92 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| SUNG METASTASIS STROMA DN | 54 | 34 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| RUAN RESPONSE TO TROGLITAZONE UP | 25 | 15 | All SZGR 2.0 genes in this pathway |
| RUAN RESPONSE TO TNF TROGLITAZONE DN | 42 | 24 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 24HR DN | 148 | 102 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR DN | 298 | 200 | All SZGR 2.0 genes in this pathway |
| RUAN RESPONSE TO TNF DN | 84 | 50 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| XU GH1 EXOGENOUS TARGETS UP | 85 | 50 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 6 | 189 | 112 | All SZGR 2.0 genes in this pathway |
| BILD CTNNB1 ONCOGENIC SIGNATURE | 82 | 52 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P7 | 90 | 52 | All SZGR 2.0 genes in this pathway |
| GAVIN IL2 RESPONSIVE FOXP3 TARGETS UP | 19 | 12 | All SZGR 2.0 genes in this pathway |
| LEE METASTASIS AND ALTERNATIVE SPLICING UP | 74 | 51 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| DE YY1 TARGETS DN | 92 | 64 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| BREDEMEYER RAG SIGNALING NOT VIA ATM DN | 57 | 34 | All SZGR 2.0 genes in this pathway |
| FIRESTEIN PROLIFERATION | 175 | 125 | All SZGR 2.0 genes in this pathway |
| ROME INSULIN TARGETS IN MUSCLE UP | 442 | 263 | All SZGR 2.0 genes in this pathway |
| COULOUARN TEMPORAL TGFB1 SIGNATURE DN | 138 | 99 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 1HR DN | 222 | 147 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE G2 | 182 | 102 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR DN | 36 | 19 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| BILANGES SERUM SENSITIVE GENES | 90 | 54 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| KATSANOU ELAVL1 TARGETS UP | 169 | 105 | All SZGR 2.0 genes in this pathway |
| JUBAN TARGETS OF SPI1 AND FLI1 UP | 115 | 73 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
| VANOEVELEN MYOGENESIS SIN3A TARGETS | 220 | 133 | All SZGR 2.0 genes in this pathway |
| BOSCO ALLERGEN INDUCED TH2 ASSOCIATED MODULE | 151 | 86 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY LUMINAL MATURE UP | 116 | 65 | All SZGR 2.0 genes in this pathway |