Gene Page: GRIK5
Summary ?
| GeneID | 2901 |
| Symbol | GRIK5 |
| Synonyms | EAA2|GRIK2|GluK5|KA2 |
| Description | glutamate ionotropic receptor kainate type subunit 5 |
| Reference | MIM:600283|HGNC:HGNC:4583|Ensembl:ENSG00000105737|HPRD:02614|Vega:OTTHUMG00000044573 |
| Gene type | protein-coding |
| Map location | 19q13.2 |
| Pascal p-value | 0.14 |
| Sherlock p-value | 0.381 |
| Fetal beta | -0.572 |
| DMG | 1 (# studies) |
| Support | CompositeSet Darnell FMRP targets Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 9 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0192 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg08955995 | 19 | 42503412 | GRIK5 | 5.273E-4 | 0.418 | 0.048 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
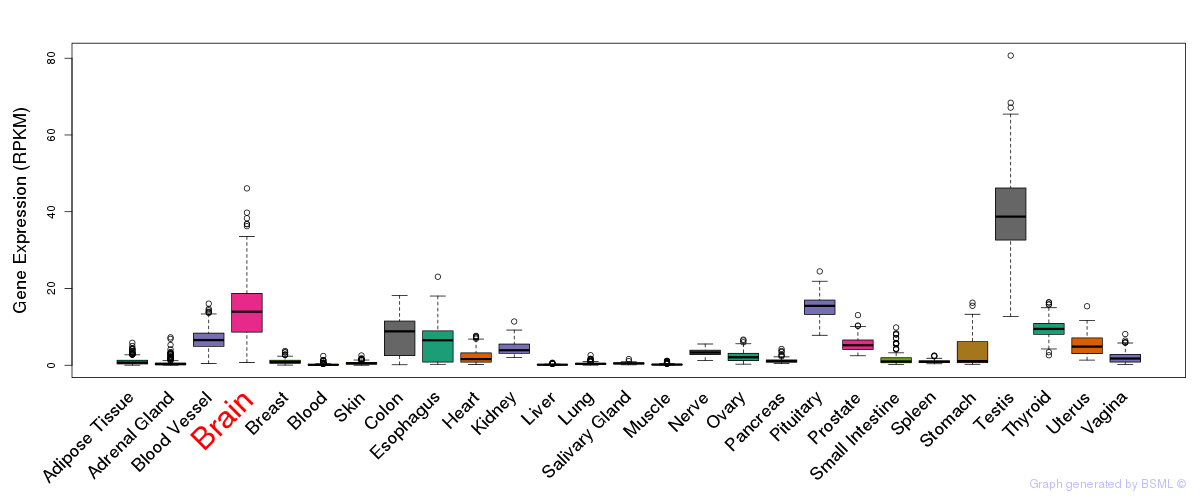
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C1orf201 | 0.80 | 0.83 |
| GRIA4 | 0.80 | 0.81 |
| STYK1 | 0.78 | 0.83 |
| LIG4 | 0.77 | 0.79 |
| GPR61 | 0.76 | 0.84 |
| WBSCR17 | 0.76 | 0.78 |
| NALCN | 0.76 | 0.84 |
| KCNC4 | 0.75 | 0.80 |
| CYB561 | 0.75 | 0.78 |
| PDGFB | 0.75 | 0.74 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SIGIRR | -0.37 | -0.45 |
| RPL18 | -0.36 | -0.46 |
| RPS16P1 | -0.36 | -0.50 |
| RPL24 | -0.36 | -0.41 |
| CSRP2 | -0.36 | -0.39 |
| RPL38 | -0.35 | -0.50 |
| UXT | -0.35 | -0.38 |
| RPL28 | -0.35 | -0.47 |
| RPL35 | -0.35 | -0.44 |
| RPL31 | -0.35 | -0.40 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005234 | extracellular-glutamate-gated ion channel activity | IEA | glutamate (GO term level: 11) | - |
| GO:0015277 | kainate selective glutamate receptor activity | IDA | glutamate (GO term level: 8) | 1321949 |
| GO:0004872 | receptor activity | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0005216 | ion channel activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0060079 | regulation of excitatory postsynaptic membrane potential | IEA | neuron, Synap (GO term level: 10) | - |
| GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane | IEA | Synap, Neurotransmitter (GO term level: 11) | - |
| GO:0007268 | synaptic transmission | NAS | neuron, Synap, Neurotransmitter (GO term level: 6) | - |
| GO:0035249 | synaptic transmission, glutamatergic | IEA | neuron, glutamate, Synap, Neurotransmitter (GO term level: 8) | - |
| GO:0006811 | ion transport | IEA | - | |
| GO:0042391 | regulation of membrane potential | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0042734 | presynaptic membrane | IEA | neuron, axon, Synap (GO term level: 5) | - |
| GO:0032983 | kainate selective glutamate receptor complex | IEA | glutamate (GO term level: 10) | - |
| GO:0045211 | postsynaptic membrane | IEA | Synap, Neurotransmitter (GO term level: 5) | - |
| GO:0045202 | synapse | IEA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0005624 | membrane fraction | IEA | - | |
| GO:0016021 | integral to membrane | NAS | - | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0030054 | cell junction | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | 186 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| REACTOME NEUROTRANSMITTER RECEPTOR BINDING AND DOWNSTREAM TRANSMISSION IN THE POSTSYNAPTIC CELL | 137 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | 31 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | 11 | 11 | All SZGR 2.0 genes in this pathway |
| GAL LEUKEMIC STEM CELL UP | 133 | 78 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL DN | 275 | 168 | All SZGR 2.0 genes in this pathway |
| YAGI AML RELAPSE PROGNOSIS | 35 | 24 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| GUO HEX TARGETS UP | 81 | 54 | All SZGR 2.0 genes in this pathway |
| YAGI AML SURVIVAL | 129 | 87 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT DN | 222 | 141 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 | 718 | 401 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC ICP WITH H3K4ME3 | 445 | 257 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA CLASSICAL | 162 | 122 | All SZGR 2.0 genes in this pathway |
| DELACROIX RARG BOUND MEF | 367 | 231 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-375 | 535 | 541 | 1A | hsa-miR-375 | UUUGUUCGUUCGGCUCGCGUGA |
| miR-376 | 449 | 455 | m8 | hsa-miR-376a | AUCAUAGAGGAAAAUCCACGU |
| hsa-miR-376b | AUCAUAGAGGAAAAUCCAUGUU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.