Gene Page: GRM8
Summary ?
| GeneID | 2918 |
| Symbol | GRM8 |
| Synonyms | GLUR8|GPRC1H|MGLUR8|mGlu8 |
| Description | glutamate receptor, metabotropic 8 |
| Reference | MIM:601116|HGNC:HGNC:4600|Ensembl:ENSG00000179603|HPRD:03071|Vega:OTTHUMG00000022888 |
| Gene type | protein-coding |
| Map location | 7q31.3-q32.1 |
| Pascal p-value | 0.018 |
| Sherlock p-value | 0.573 |
| Fetal beta | -1.686 |
| DMG | 1 (# studies) |
| eGene | Cerebellum |
| Support | GPCR SIGNALLING Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| CV:PheWAS | Phenome-wide association studies (PheWAS) | 157 SNPs associated with schizophrenia | 1 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 1 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 CV:PheWAS
CV:PheWAS
| SNP ID | Chromosome | Position | Allele | P | Function | Gene | Up/Down Distance |
|---|---|---|---|---|---|---|---|
| rs17864092 | 7 | 126639604 | null | 0.4378 | GRM8 | null |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg02409125 | 7 | 126889555 | GRM8 | 1.881E-4 | 0.337 | 0.034 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
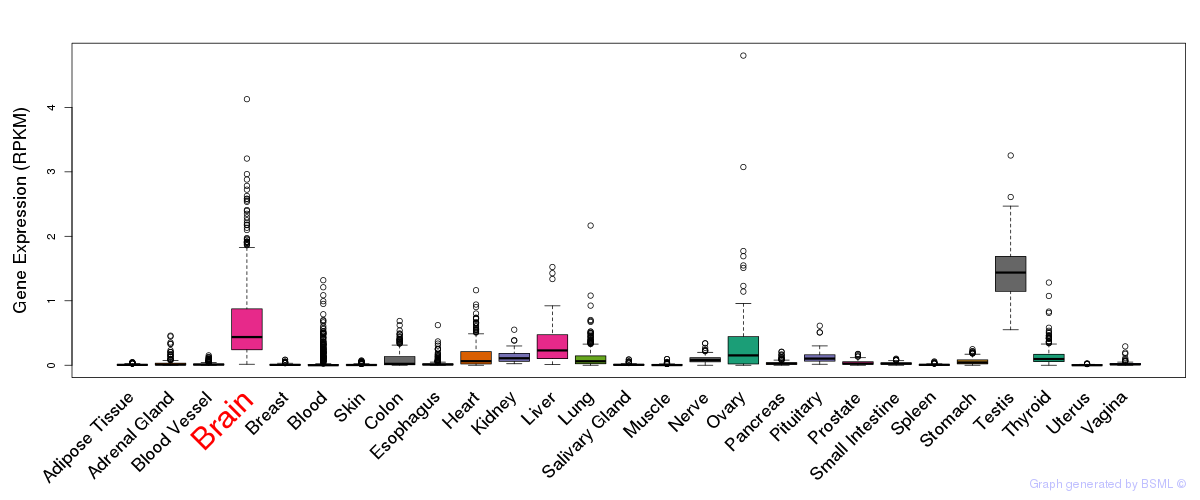
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PPP2R5E | 0.97 | 0.97 |
| DIS3 | 0.96 | 0.97 |
| ZMYM4 | 0.96 | 0.97 |
| WDR48 | 0.96 | 0.97 |
| NUP133 | 0.96 | 0.97 |
| WDR3 | 0.96 | 0.97 |
| SUPT16H | 0.95 | 0.96 |
| EXOC2 | 0.95 | 0.97 |
| STRN3 | 0.95 | 0.97 |
| TAF2 | 0.95 | 0.96 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.78 | -0.88 |
| MT-CO2 | -0.78 | -0.89 |
| FXYD1 | -0.76 | -0.86 |
| AF347015.27 | -0.76 | -0.85 |
| AF347015.33 | -0.76 | -0.85 |
| AF347015.8 | -0.75 | -0.87 |
| MT-CYB | -0.75 | -0.85 |
| IFI27 | -0.74 | -0.85 |
| HIGD1B | -0.73 | -0.85 |
| AF347015.15 | -0.72 | -0.85 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0001642 | group III metabotropic glutamate receptor activity | IEA | glutamate (GO term level: 8) | - |
| GO:0004872 | receptor activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007186 | G-protein coupled receptor protein signaling pathway | IEA | - | |
| GO:0007194 | negative regulation of adenylate cyclase activity | TAS | 9473604 | |
| GO:0007165 | signal transduction | IEA | - | |
| GO:0007608 | sensory perception of smell | IEA | - | |
| GO:0007601 | visual perception | TAS | 9473604 | |
| GO:0050896 | response to stimulus | IEA | - | |
| GO:0030818 | negative regulation of cAMP biosynthetic process | IDA | 11166323 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 9473604 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | 15 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR LIGAND BINDING | 408 | 246 | All SZGR 2.0 genes in this pathway |
| WATANABE COLON CANCER MSI VS MSS DN | 81 | 42 | All SZGR 2.0 genes in this pathway |
| SABATES COLORECTAL ADENOMA UP | 141 | 75 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 6HR DN | 160 | 101 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K27ME3 UP | 295 | 149 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME3 AND H3K27ME3 | 142 | 103 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION | 368 | 247 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS WITH HCP H3K27ME3 | 102 | 76 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-219 | 97 | 103 | 1A | hsa-miR-219brain | UGAUUGUCCAAACGCAAUUCU |
| miR-24 | 146 | 152 | 1A | hsa-miR-24SZ | UGGCUCAGUUCAGCAGGAACAG |
| miR-33 | 446 | 452 | 1A | hsa-miR-33 | GUGCAUUGUAGUUGCAUUG |
| hsa-miR-33b | GUGCAUUGCUGUUGCAUUGCA | ||||
| miR-339 | 2 | 8 | 1A | hsa-miR-339 | UCCCUGUCCUCCAGGAGCUCA |
| miR-342 | 340 | 346 | 1A | hsa-miR-342brain | UCUCACACAGAAAUCGCACCCGUC |
| miR-381 | 375 | 382 | 1A,m8 | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| miR-543 | 504 | 510 | m8 | hsa-miR-543 | AAACAUUCGCGGUGCACUUCU |
| hsa-miR-543 | AAACAUUCGCGGUGCACUUCU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.