Gene Page: KCNJ8
Summary ?
| GeneID | 3764 |
| Symbol | KCNJ8 |
| Synonyms | KIR6.1|uKATP-1 |
| Description | potassium voltage-gated channel subfamily J member 8 |
| Reference | MIM:600935|HGNC:HGNC:6269|Ensembl:ENSG00000121361|HPRD:02962|Vega:OTTHUMG00000169093 |
| Gene type | protein-coding |
| Map location | 12p11.23 |
| Pascal p-value | 0.47 |
| Sherlock p-value | 0.205 |
| Fetal beta | -1.182 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
| Support | EXCITABILITY |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 1 |
| Expression | Meta-analysis of gene expression | P value: 1.517 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg03243946 | 12 | 21811034 | KCNJ8 | 0.004 | -2.405 | DMG:vanEijk_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs11046386 | chr12 | 22513129 | KCNJ8 | 3764 | 0.16 | cis | ||
| rs12316081 | chr12 | 22515892 | KCNJ8 | 3764 | 0.1 | cis | ||
| rs10841998 | chr12 | 22531239 | KCNJ8 | 3764 | 0.08 | cis | ||
| rs12303966 | chr12 | 22665916 | KCNJ8 | 3764 | 0.2 | cis |
Section II. Transcriptome annotation
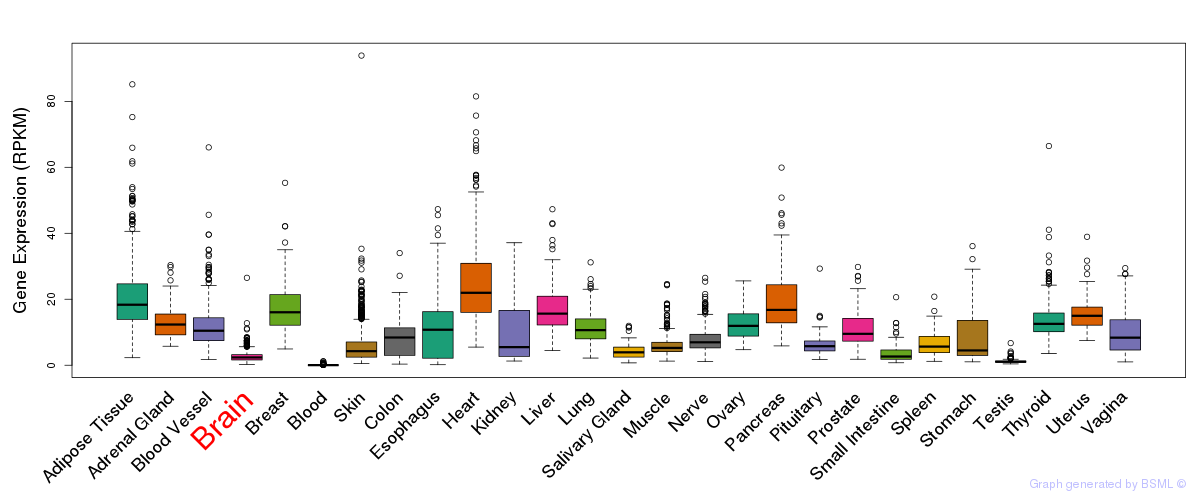
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005244 | voltage-gated ion channel activity | IEA | - | |
| GO:0015272 | ATP-activated inward rectifier potassium channel activity | IEA | - | |
| GO:0030955 | potassium ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007507 | heart development | IEA | - | |
| GO:0006811 | ion transport | IEA | - | |
| GO:0006813 | potassium ion transport | TAS | 7890693 | |
| GO:0051607 | defense response to virus | IEA | - | |
| GO:0032496 | response to lipopolysaccharide | IEA | - | |
| GO:0043330 | response to exogenous dsRNA | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0030016 | myofibril | IEA | - | |
| GO:0008076 | voltage-gated potassium channel complex | TAS | 7890693 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| REACTOME POTASSIUM CHANNELS | 98 | 68 | All SZGR 2.0 genes in this pathway |
| REACTOME INWARDLY RECTIFYING K CHANNELS | 31 | 20 | All SZGR 2.0 genes in this pathway |
| LIU PROSTATE CANCER DN | 481 | 290 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE DN | 244 | 147 | All SZGR 2.0 genes in this pathway |
| ROY WOUND BLOOD VESSEL UP | 50 | 30 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| ROVERSI GLIOMA LOH REGIONS | 44 | 30 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 5 | 147 | 89 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM2 | 153 | 102 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| IIZUKA LIVER CANCER PROGRESSION G1 G2 UP | 12 | 9 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| CHEN LVAD SUPPORT OF FAILING HEART DN | 42 | 32 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE DN | 261 | 183 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS CTNNB1 UP | 176 | 110 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS PROLIFERATION DN | 179 | 97 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 | 491 | 319 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S3 | 266 | 180 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |