Gene Page: ARF6
Summary ?
| GeneID | 382 |
| Symbol | ARF6 |
| Synonyms | - |
| Description | ADP ribosylation factor 6 |
| Reference | MIM:600464|HGNC:HGNC:659|Ensembl:ENSG00000165527|HPRD:02714|Vega:OTTHUMG00000140296 |
| Gene type | protein-coding |
| Map location | 14q21.3 |
| Pascal p-value | 0.627 |
| Fetal beta | -0.064 |
| eGene | Cerebellar Hemisphere Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0192 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs11682393 | chr2 | 47804496 | ARF6 | 382 | 0.1 | trans |
Section II. Transcriptome annotation
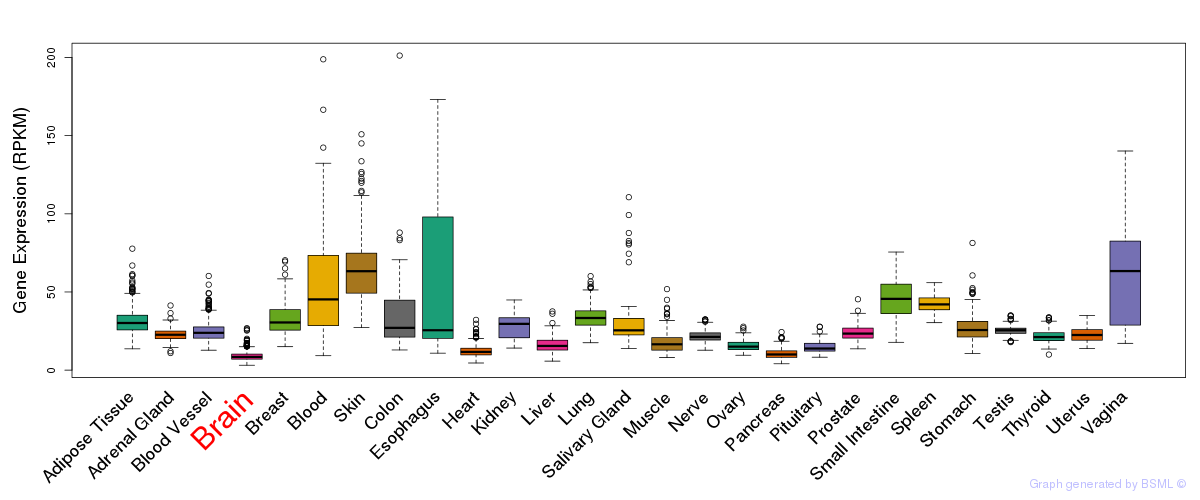
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0003924 | GTPase activity | TAS | 1993656 | |
| GO:0005515 | protein binding | IPI | 9312003 | |
| GO:0005525 | GTP binding | TAS | 10913182 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001889 | liver development | IEA | - | |
| GO:0007155 | cell adhesion | TAS | 10036235 | |
| GO:0006928 | cell motion | TAS | 10036235 | |
| GO:0007264 | small GTPase mediated signal transduction | IEA | - | |
| GO:0006915 | apoptosis | IEA | - | |
| GO:0016192 | vesicle-mediated transport | TAS | 10913182 | |
| GO:0035020 | regulation of Rac protein signal transduction | IDA | 10036235 | |
| GO:0015031 | protein transport | IEA | - | |
| GO:0031529 | ruffle organization | IDA | 10036235 | |
| GO:0048261 | negative regulation of receptor-mediated endocytosis | TAS | 9312003 | |
| GO:0030838 | positive regulation of actin filament polymerization | IMP | 10036235 | |
| GO:0030866 | cortical actin cytoskeleton organization | IMP | 10913182 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001726 | ruffle | IDA | 9312003 | |
| GO:0005794 | Golgi apparatus | IEA | - | |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005624 | membrane fraction | TAS | 8702973 | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005768 | endosome | TAS | 10913182 | |
| GO:0005938 | cell cortex | IDA | 10913182 | |
| GO:0005886 | plasma membrane | IDA | 10036235 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AP1B1 | ADTB1 | AP105A | BAM22 | CLAPB2 | adaptor-related protein complex 1, beta 1 subunit | - | HPRD | 11926829 |
| AP3B1 | ADTB3 | ADTB3A | HPS | HPS2 | PE | adaptor-related protein complex 3, beta 1 subunit | Reconstituted Complex | BioGRID | 11926829 |
| AP3D1 | ADTD | hBLVR | adaptor-related protein complex 3, delta 1 subunit | - | HPRD | 11926829 |
| AP3S2 | AP3S3 | FLJ35955 | sigma3b | adaptor-related protein complex 3, sigma 2 subunit | Reconstituted Complex | BioGRID | 11926829 |
| ARFIP2 | POR1 | ADP-ribosylation factor interacting protein 2 | - | HPRD,BioGRID | 9312003 |
| ARRB1 | ARB1 | ARR1 | arrestin, beta 1 | - | HPRD,BioGRID | 11533043 |11867621 |
| ARRB2 | ARB2 | ARR2 | BARR2 | DKFZp686L0365 | arrestin, beta 2 | - | HPRD,BioGRID | 11533043 |11867621 |
| ASAP2 | AMAP2 | CENTB3 | DDEF2 | FLJ42910 | KIAA0400 | PAG3 | PAP | Pap-alpha | SHAG1 | ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 | - | HPRD | 10022920 |10749932 |
| CHRM3 | HM3 | cholinergic receptor, muscarinic 3 | - | HPRD,BioGRID | 12799371 |
| CYTH2 | ARNO | CTS18 | CTS18.1 | PSCD2 | PSCD2L | SEC7L | Sec7p-L | Sec7p-like | cytohesin 2 | - | HPRD,BioGRID | 9417041 |
| DHPS | MIG13 | deoxyhypusine synthase | Affinity Capture-MS | BioGRID | 17353931 |
| DYNC1H1 | DHC1 | DHC1a | DKFZp686P2245 | DNCH1 | DNCL | DNECL | DYHC | Dnchc1 | HL-3 | KIAA0325 | p22 | dynein, cytoplasmic 1, heavy chain 1 | Affinity Capture-MS | BioGRID | 17353931 |
| EIF6 | 2 | CAB | EIF3A | ITGB4BP | b | b(2)gcn | gcn | p27BBP | eukaryotic translation initiation factor 6 | Affinity Capture-MS | BioGRID | 17353931 |
| EPPK1 | EPIPL | EPIPL1 | epiplakin 1 | Affinity Capture-MS | BioGRID | 17353931 |
| EXOC5 | DKFZp666H126 | HSEC10 | PRO1912 | SEC10 | SEC10L1 | SEC10P | exocyst complex component 5 | - | HPRD,BioGRID | 14662749 |
| GART | AIRS | GARS | GARTF | MGC47764 | PAIS | PGFT | PRGS | phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase | Affinity Capture-MS | BioGRID | 17353931 |
| IQGAP1 | HUMORFA01 | KIAA0051 | SAR1 | p195 | IQ motif containing GTPase activating protein 1 | Affinity Capture-MS | BioGRID | 17353931 |
| KIAA0368 | ECM29 | FLJ22036 | KIAA1962 | RP11-386D8.2 | KIAA0368 | Affinity Capture-MS | BioGRID | 17353931 |
| LRPPRC | CLONE-23970 | GP130 | LRP130 | LSFC | leucine-rich PPR-motif containing | Affinity Capture-MS | BioGRID | 17353931 |
| PABPC4 | APP-1 | APP1 | FLJ43938 | PABP4 | iPABP | poly(A) binding protein, cytoplasmic 4 (inducible form) | Affinity Capture-MS | BioGRID | 17353931 |
| PIP5K1A | - | phosphatidylinositol-4-phosphate 5-kinase, type I, alpha | - | HPRD | 10589680 |
| PIP5K1C | KIAA0589 | LCCS3 | PIP5K-GAMMA | PIP5Kgamma | phosphatidylinositol-4-phosphate 5-kinase, type I, gamma | - | HPRD,BioGRID | 12847086 |
| PLD1 | - | phospholipase D1, phosphatidylcholine-specific | - | HPRD,BioGRID | 12379803 |
| RAB11FIP3 | KIAA0665 | Rab11-FIP3 | RAB11 family interacting protein 3 (class II) | - | HPRD | 11535061 |
| RALA | MGC48949 | RAL | v-ral simian leukemia viral oncogene homolog A (ras related) | Affinity Capture-Western | BioGRID | 12509462 |
| YARS | CMTDIC | TYRRS | YRS | YTS | tyrosyl-tRNA synthetase | Affinity Capture-MS | BioGRID | 17353931 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ENDOCYTOSIS | 183 | 132 | All SZGR 2.0 genes in this pathway |
| KEGG FC GAMMA R MEDIATED PHAGOCYTOSIS | 97 | 71 | All SZGR 2.0 genes in this pathway |
| PID MET PATHWAY | 80 | 60 | All SZGR 2.0 genes in this pathway |
| PID ARF6 TRAFFICKING PATHWAY | 49 | 34 | All SZGR 2.0 genes in this pathway |
| PID ARF6 PATHWAY | 35 | 27 | All SZGR 2.0 genes in this pathway |
| PID ARF6 DOWNSTREAM PATHWAY | 15 | 14 | All SZGR 2.0 genes in this pathway |
| PID PI3KCI PATHWAY | 49 | 40 | All SZGR 2.0 genes in this pathway |
| PID AJDISS 2PATHWAY | 48 | 38 | All SZGR 2.0 genes in this pathway |
| PID ECADHERIN NASCENT AJ PATHWAY | 39 | 33 | All SZGR 2.0 genes in this pathway |
| PID ECADHERIN STABILIZATION PATHWAY | 42 | 34 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN A4B1 PATHWAY | 33 | 24 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE UP | 423 | 283 | All SZGR 2.0 genes in this pathway |
| HORIUCHI WTAP TARGETS DN | 310 | 188 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM1 | 229 | 137 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| SCIAN INVERSED TARGETS OF TP53 AND TP73 DN | 31 | 23 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 240 HELA | 60 | 43 | All SZGR 2.0 genes in this pathway |
| SUNG METASTASIS STROMA DN | 54 | 34 | All SZGR 2.0 genes in this pathway |
| GOLDRATH HOMEOSTATIC PROLIFERATION | 171 | 102 | All SZGR 2.0 genes in this pathway |
| CHEOK RESPONSE TO HD MTX DN | 24 | 18 | All SZGR 2.0 genes in this pathway |
| FLECHNER PBL KIDNEY TRANSPLANT REJECTED VS OK UP | 63 | 48 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| LEI MYB TARGETS | 318 | 215 | All SZGR 2.0 genes in this pathway |
| XU CREBBP TARGETS DN | 44 | 31 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D4 | 55 | 37 | All SZGR 2.0 genes in this pathway |
| JI RESPONSE TO FSH DN | 58 | 43 | All SZGR 2.0 genes in this pathway |
| GENTILE UV HIGH DOSE DN | 312 | 203 | All SZGR 2.0 genes in this pathway |
| MCCLUNG CREB1 TARGETS UP | 100 | 72 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS UP | 198 | 132 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| HILLION HMGA1B TARGETS | 92 | 68 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN UP | 439 | 257 | All SZGR 2.0 genes in this pathway |
| LABBE TARGETS OF TGFB1 AND WNT3A DN | 108 | 68 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| HINATA NFKB TARGETS FIBROBLAST UP | 84 | 60 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS UP | 518 | 299 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH INV 16 TRANSLOCATION | 422 | 277 | All SZGR 2.0 genes in this pathway |
| LI INDUCED T TO NATURAL KILLER UP | 307 | 182 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 3 DN | 42 | 24 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 UP | 344 | 215 | All SZGR 2.0 genes in this pathway |
| HOLLEMAN ASPARAGINASE RESISTANCE B ALL DN | 15 | 10 | All SZGR 2.0 genes in this pathway |
| HOLLEMAN ASPARAGINASE RESISTANCE ALL DN | 25 | 14 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-139 | 425 | 431 | 1A | hsa-miR-139brain | UCUACAGUGCACGUGUCU |
| miR-23 | 577 | 583 | m8 | hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC |
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| miR-30-3p | 485 | 492 | 1A,m8 | hsa-miR-30a-3p | CUUUCAGUCGGAUGUUUGCAGC |
| hsa-miR-30e-3p | CUUUCAGUCGGAUGUUUACAGC | ||||
| miR-323 | 577 | 583 | 1A | hsa-miR-323brain | GCACAUUACACGGUCGACCUCU |
| miR-494 | 470 | 476 | 1A | hsa-miR-494brain | UGAAACAUACACGGGAAACCUCUU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.