Gene Page: LRP2
Summary ?
| GeneID | 4036 |
| Symbol | LRP2 |
| Synonyms | DBS|GP330 |
| Description | LDL receptor related protein 2 |
| Reference | MIM:600073|HGNC:HGNC:6694|Ensembl:ENSG00000081479|HPRD:02509|Vega:OTTHUMG00000132179 |
| Gene type | protein-coding |
| Map location | 2q31.1 |
| Pascal p-value | 0.273 |
| Fetal beta | -1.022 |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.02395 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 1.3965 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
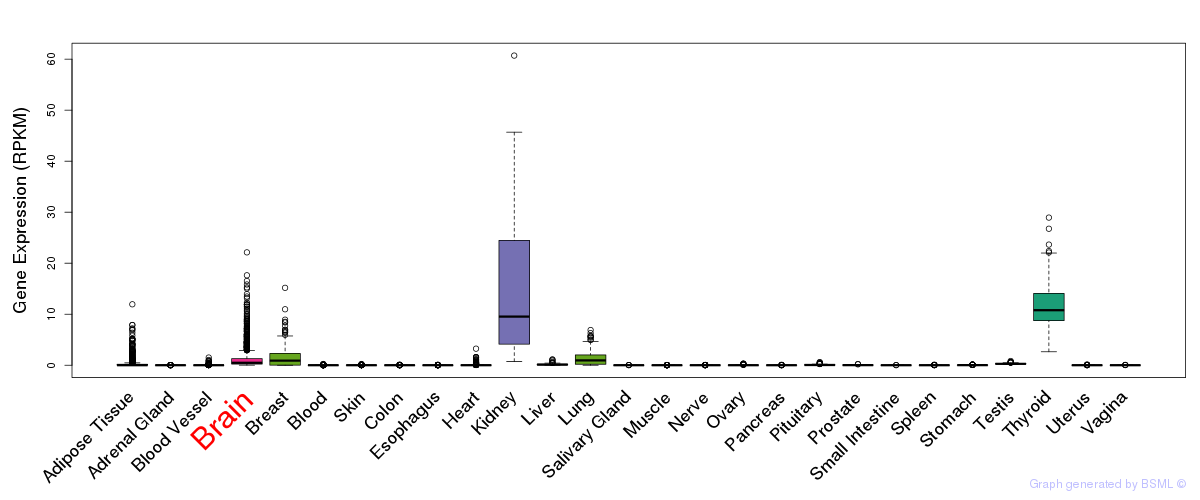
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RUNDC3A | 0.87 | 0.88 |
| HAGH | 0.86 | 0.88 |
| RHBDD2 | 0.86 | 0.88 |
| PGP | 0.84 | 0.87 |
| FKBP8 | 0.84 | 0.88 |
| ABHD8 | 0.83 | 0.87 |
| NBL1 | 0.83 | 0.81 |
| RAB26 | 0.83 | 0.81 |
| PEF1 | 0.83 | 0.82 |
| STUB1 | 0.82 | 0.88 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| EIF5B | -0.58 | -0.68 |
| THOC2 | -0.54 | -0.52 |
| AC005921.3 | -0.52 | -0.74 |
| CCAR1 | -0.51 | -0.48 |
| ZNF326 | -0.51 | -0.51 |
| UPF3B | -0.50 | -0.53 |
| SFRS12 | -0.50 | -0.48 |
| RBM25 | -0.49 | -0.50 |
| MAP4K4 | -0.48 | -0.46 |
| SFRS18 | -0.48 | -0.63 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004872 | receptor activity | IEA | - | |
| GO:0005529 | sugar binding | IEA | - | |
| GO:0005509 | calcium ion binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 9773776 |14528014 | |
| GO:0017124 | SH3 domain binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0030900 | forebrain development | IEA | Brain (GO term level: 8) | - |
| GO:0006486 | protein amino acid glycosylation | TAS | 2786251 | |
| GO:0008283 | cell proliferation | IEA | - | |
| GO:0006766 | vitamin metabolic process | IEA | - | |
| GO:0006629 | lipid metabolic process | TAS | 7768901 | |
| GO:0006898 | receptor-mediated endocytosis | TAS | 7768901 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005794 | Golgi apparatus | IEA | - | |
| GO:0005624 | membrane fraction | IEA | - | |
| GO:0005764 | lysosome | TAS | 7768901 | |
| GO:0005768 | endosome | IEA | - | |
| GO:0005783 | endoplasmic reticulum | IEA | - | |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005905 | coated pit | IEA | - | |
| GO:0005903 | brush border | IEA | - | |
| GO:0016324 | apical plasma membrane | IEA | - | |
| GO:0030139 | endocytic vesicle | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ALB | DKFZp779N1935 | PRO0883 | PRO0903 | PRO1341 | albumin | - | HPRD | 8898021|11158855 |
| ANAPC10 | APC10 | DKFZp564L0562 | DOC1 | anaphase promoting complex subunit 10 | Two-hybrid | BioGRID | 10827173 |
| ANKRA2 | ANKRA | ankyrin repeat, family A (RFXANK-like), 2 | - | HPRD,BioGRID | 11095640 |
| ANKS1B | AIDA | AIDA-1 | ANKS2 | EB-1 | MGC26087 | cajalin-2 | ankyrin repeat and sterile alpha motif domain containing 1B | - | HPRD,BioGRID | 12508107 |
| APBA2 | D15S1518E | HsT16821 | LIN-10 | MGC99508 | MGC:14091 | MINT2 | X11L | amyloid beta (A4) precursor protein-binding, family A, member 2 | Two-hybrid | BioGRID | 10827173 |
| APBB1 | FE65 | MGC:9072 | RIR | amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) | - | HPRD | 9837937 |
| APOB | FLDB | apolipoprotein B (including Ag(x) antigen) | - | HPRD | 10330424 |
| APOE | AD2 | LPG | MGC1571 | apoprotein | apolipoprotein E | - | HPRD | 11421580 |
| APOE | AD2 | LPG | MGC1571 | apoprotein | apolipoprotein E | Reconstituted Complex | BioGRID | 7768901 |
| APOH | B2G1 | BG | apolipoprotein H (beta-2-glycoprotein I) | - | HPRD,BioGRID | 9727058 |
| CLU | AAG4 | APOJ | CLI | KUB1 | MGC24903 | SGP-2 | SGP2 | SP-40 | TRPM-2 | TRPM2 | clusterin | - | HPRD,BioGRID | 7768901 |
| CUBN | FLJ90055 | FLJ90747 | IFCR | MGA1 | gp280 | cubilin (intrinsic factor-cobalamin receptor) | - | HPRD | 11595644|11994745 |
| DAB1 | - | disabled homolog 1 (Drosophila) | - | HPRD,BioGRID | 10827173 |
| DAB2 | DOC-2 | DOC2 | FLJ26626 | disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila) | - | HPRD,BioGRID | 10769163 |
| DLG1 | DKFZp761P0818 | DKFZp781B0426 | DLGH1 | SAP97 | dJ1061C18.1.1 | hdlg | discs, large homolog 1 (Drosophila) | - | HPRD | 12713445 |
| DLG2 | DKFZp781D1854 | DKFZp781E0954 | FLJ37266 | MGC131811 | PSD-93 | discs, large homolog 2, chapsyn-110 (Drosophila) | - | HPRD,BioGRID | 12713445 |
| DLG3 | KIAA1232 | MRX | MRX90 | NE-Dlg | NEDLG | SAP102 | discs, large homolog 3 (neuroendocrine-dlg, Drosophila) | - | HPRD,BioGRID | 12713445 |
| DLG4 | FLJ97752 | FLJ98574 | PSD95 | SAP90 | discs, large homolog 4 (Drosophila) | - | HPRD,BioGRID | 12713445 |
| GC | DBP | DBP/GC | VDBG | VDBP | group-specific component (vitamin D binding protein) | - | HPRD,BioGRID | 11717447 |
| GIPC1 | C19orf3 | GIPC | GLUT1CBP | Hs.6454 | IIP-1 | MGC15889 | MGC3774 | NIP | RGS19IP1 | SEMCAP | SYNECTIIN | SYNECTIN | TIP-2 | GIPC PDZ domain containing family, member 1 | - | HPRD,BioGRID | 11912251 |
| GPRASP1 | GASP | GASP1 | KIAA0443 | G protein-coupled receptor associated sorting protein 1 | - | HPRD | 11274227 |
| INS | ILPR | IRDN | insulin | - | HPRD | 9773776 |
| ITGB1BP1 | DKFZp686K08158 | ICAP-1A | ICAP-1B | ICAP-1alpha | ICAP1 | ICAP1A | ICAP1B | integrin beta 1 binding protein 1 | - | HPRD,BioGRID | 10827173 |
| LDLRAP1 | ARH | ARH1 | ARH2 | DKFZp586D0624 | FHCB1 | FHCB2 | MGC34705 | low density lipoprotein receptor adaptor protein 1 | - | HPRD,BioGRID | 14528014 |
| LPA | AK38 | APOA | LP | lipoprotein, Lp(a) | - | HPRD,BioGRID | 10073957 |
| LPL | HDLCQ11 | LIPD | lipoprotein lipase | - | HPRD,BioGRID | 7686151 |
| LRP2BP | DKFZp761O0113 | FLJ44965 | LRP2 binding protein | - | HPRD,BioGRID | 12508107 |
| LRPAP1 | A2MRAP | A2RAP | HBP44 | MGC138272 | MRAP | RAP | low density lipoprotein receptor-related protein associated protein 1 | - | HPRD,BioGRID | 7512726 |
| MAGI1 | AIP3 | BAIAP1 | BAP1 | MAGI-1 | TNRC19 | WWP3 | membrane associated guanylate kinase, WW and PDZ domain containing 1 | - | HPRD,BioGRID | 11274227 |
| MAPK8IP1 | IB1 | JIP-1 | JIP1 | PRKM8IP | mitogen-activated protein kinase 8 interacting protein 1 | - | HPRD,BioGRID | 10827173 |
| MAPK8IP2 | IB2 | JIP2 | PRKM8IPL | mitogen-activated protein kinase 8 interacting protein 2 | Reconstituted Complex Two-hybrid | BioGRID | 10827173 |12508107 |
| MDK | FLJ27379 | MK | NEGF2 | midkine (neurite growth-promoting factor 2) | - | HPRD | 10772929 |
| NOS1AP | 6330408P19Rik | CAPON | MGC138500 | nitric oxide synthase 1 (neuronal) adaptor protein | - | HPRD,BioGRID | 10827173 |
| PLAU | ATF | UPA | URK | UROKINASE | u-PA | plasminogen activator, urokinase | - | HPRD,BioGRID | 8344937 |
| RBP1 | CRABP-I | CRBP | CRBP1 | CRBPI | RBPC | retinol binding protein 1, cellular | - | HPRD,BioGRID | 10203351 |
| SCGB1A1 | CC10 | CC16 | CCSP | UGB | secretoglobin, family 1A, member 1 (uteroglobin) | - | HPRD,BioGRID | 11278724 |
| SCN3A | KIAA1356 | NAC3 | Nav1.3 | sodium channel, voltage-gated, type III, alpha subunit | Two-hybrid | BioGRID | 10827173 |
| SERPINE1 | PAI | PAI-1 | PAI1 | PLANH1 | serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 | - | HPRD | 8344937 |
| SLC9A3 | MGC126718 | MGC126720 | NHE3 | solute carrier family 9 (sodium/hydrogen exchanger), member 3 | - | HPRD | 10364184 |
| SNX17 | KIAA0064 | sorting nexin 17 | Reconstituted Complex | BioGRID | 12169628 |
| SYNJ2BP | ARIP2 | FLJ11271 | FLJ41973 | OMP25 | synaptojanin 2 binding protein | - | HPRD,BioGRID | 10827173 |
| TG | AITD3 | TGN | thyroglobulin | - | HPRD | 9492085 |
| THBS1 | THBS | TSP | TSP1 | thrombospondin 1 | Reconstituted Complex | BioGRID | 7775583 |
| TLN1 | ILWEQ | KIAA1027 | TLN | talin 1 | - | HPRD | 10827173 |
| TTR | HsT2651 | PALB | TBPA | transthyretin | - | HPRD | 10982792 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG HEDGEHOG SIGNALING PATHWAY | 56 | 42 | All SZGR 2.0 genes in this pathway |
| PID HEDGEHOG 2PATHWAY | 22 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | 35 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME STEROID HORMONES | 29 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| GAL LEUKEMIC STEM CELL DN | 244 | 153 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER DN | 232 | 154 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| BECKER TAMOXIFEN RESISTANCE DN | 52 | 37 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| AFFAR YY1 TARGETS UP | 214 | 133 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| LOPEZ EPITHELIOID MESOTHELIOMA | 17 | 11 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 11 | 103 | 68 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-101 | 1069 | 1076 | 1A,m8 | hsa-miR-101 | UACAGUACUGUGAUAACUGAAG |
| miR-139 | 560 | 567 | 1A,m8 | hsa-miR-139brain | UCUACAGUGCACGUGUCU |
| miR-142-5p | 86 | 92 | m8 | hsa-miR-142-5p | CAUAAAGUAGAAAGCACUAC |
| miR-144 | 1070 | 1076 | 1A | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| miR-15/16/195/424/497 | 1441 | 1447 | m8 | hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG |
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| miR-19 | 38 | 45 | 1A,m8 | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-204/211 | 1000 | 1007 | 1A,m8 | hsa-miR-204brain | UUCCCUUUGUCAUCCUAUGCCU |
| hsa-miR-211 | UUCCCUUUGUCAUCCUUCGCCU | ||||
| miR-409-3p | 76 | 83 | 1A,m8 | hsa-miR-409-3p | CGAAUGUUGCUCGGUGAACCCCU |
| miR-455 | 41 | 47 | 1A | hsa-miR-455 | UAUGUGCCUUUGGACUACAUCG |
| miR-93.hd/291-3p/294/295/302/372/373/520 | 469 | 476 | 1A,m8 | hsa-miR-93brain | AAAGUGCUGUUCGUGCAGGUAG |
| hsa-miR-302a | UAAGUGCUUCCAUGUUUUGGUGA | ||||
| hsa-miR-302b | UAAGUGCUUCCAUGUUUUAGUAG | ||||
| hsa-miR-302c | UAAGUGCUUCCAUGUUUCAGUGG | ||||
| hsa-miR-302d | UAAGUGCUUCCAUGUUUGAGUGU | ||||
| hsa-miR-372 | AAAGUGCUGCGACAUUUGAGCGU | ||||
| hsa-miR-373 | GAAGUGCUUCGAUUUUGGGGUGU | ||||
| hsa-miR-520e | AAAGUGCUUCCUUUUUGAGGG | ||||
| hsa-miR-520a | AAAGUGCUUCCCUUUGGACUGU | ||||
| hsa-miR-520b | AAAGUGCUUCCUUUUAGAGGG | ||||
| hsa-miR-520c | AAAGUGCUUCCUUUUAGAGGGUU | ||||
| hsa-miR-520d | AAAGUGCUUCUCUUUGGUGGGUU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.