Gene Page: LRPAP1
Summary ?
| GeneID | 4043 |
| Symbol | LRPAP1 |
| Synonyms | A2MRAP|A2RAP|HBP44|MRAP|MYP23|RAP|alpha-2-MRAP |
| Description | LDL receptor related protein associated protein 1 |
| Reference | MIM:104225|HGNC:HGNC:6701|HPRD:00082| |
| Gene type | protein-coding |
| Map location | 4p16.3 |
| Pascal p-value | 0.437 |
| Sherlock p-value | 0.555 |
| Fetal beta | -1.606 |
| eGene | Cerebellar Hemisphere Cerebellum Frontal Cortex BA9 Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
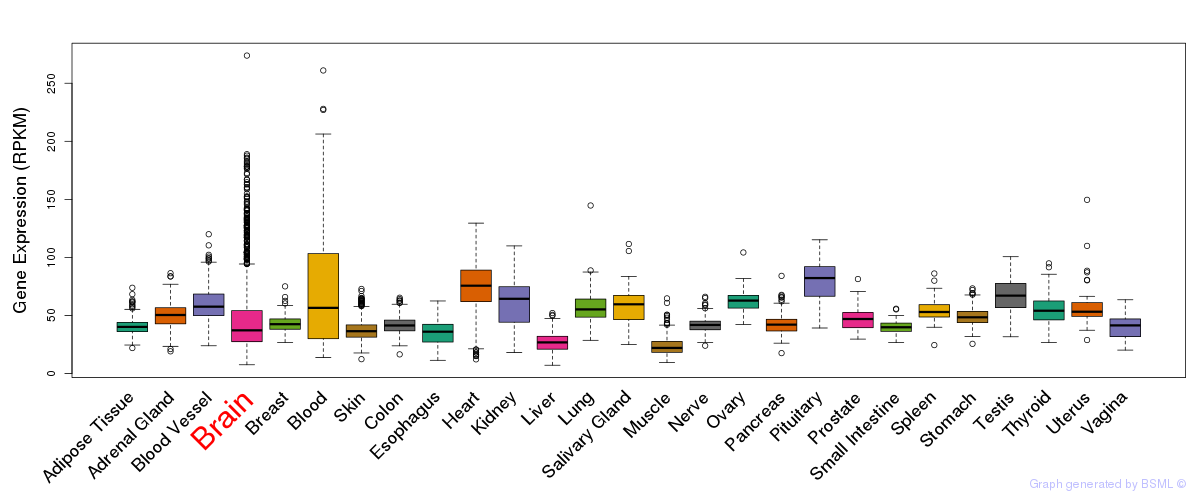
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| EIF4EBP1 | 4E-BP1 | 4EBP1 | BP-1 | MGC4316 | PHAS-I | eukaryotic translation initiation factor 4E binding protein 1 | Affinity Capture-Western | BioGRID | 16798736 |
| FRAP1 | FLJ44809 | FRAP | FRAP2 | MTOR | RAFT1 | RAPT1 | FK506 binding protein 12-rapamycin associated protein 1 | Affinity Capture-Western | BioGRID | 16798736 |
| LDLR | FH | FHC | low density lipoprotein receptor | - | HPRD,BioGRID | 7822276 |
| LRP1 | A2MR | APOER | APR | CD91 | FLJ16451 | IGFBP3R | LRP | MGC88725 | TGFBR5 | low density lipoprotein-related protein 1 (alpha-2-macroglobulin receptor) | - | HPRD,BioGRID | 1374383 |1718973 |10747918 |
| LRP1B | LRP-DIT | LRPDIT | low density lipoprotein-related protein 1B (deleted in tumors) | - | HPRD | 11384978 |
| LRP2 | DBS | gp330 | low density lipoprotein-related protein 2 | - | HPRD,BioGRID | 7512726 |
| LRP8 | APOER2 | HSZ75190 | MCI1 | low density lipoprotein receptor-related protein 8, apolipoprotein e receptor | - | HPRD,BioGRID | 12899622 |
| POLA2 | FLJ21662 | FLJ37250 | polymerase (DNA directed), alpha 2 (70kD subunit) | Two-hybrid | BioGRID | 16169070 |
| RAF1 | CRAF | NS5 | Raf-1 | c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 | Two-hybrid | BioGRID | 10783161 |
| RALA | MGC48949 | RAL | v-ral simian leukemia viral oncogene homolog A (ras related) | Reconstituted Complex | BioGRID | 11167825 |
| RALGDS | FLJ20922 | RGF | RalGEF | ral guanine nucleotide dissociation stimulator | Two-hybrid | BioGRID | 10783161 |
| RTN4 | ASY | NI220/250 | NOGO | NOGO-A | NOGOC | NSP | NSP-CL | Nbla00271 | Nbla10545 | Nogo-B | Nogo-C | RTN-X | RTN4-A | RTN4-B1 | RTN4-B2 | RTN4-C | reticulon 4 | Two-hybrid | BioGRID | 16169070 |
| SORL1 | C11orf32 | FLJ21930 | FLJ39258 | LR11 | LRP9 | SORLA | SorLA-1 | gp250 | sortilin-related receptor, L(DLR class) A repeats-containing | - | HPRD,BioGRID | 8940146 |11557679 |
| SORT1 | Gp95 | NT3 | sortilin 1 | Reconstituted Complex | BioGRID | 9013611 |
| VLDLR | CHRMQ1 | FLJ35024 | VLDLRCH | very low density lipoprotein receptor | - | HPRD | 7592875 |10571241 |
| VLDLR | CHRMQ1 | FLJ35024 | VLDLRCH | very low density lipoprotein receptor | Reconstituted Complex | BioGRID | 10571241 |12899622 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID REELIN PATHWAY | 29 | 29 | All SZGR 2.0 genes in this pathway |
| PID LIS1 PATHWAY | 28 | 22 | All SZGR 2.0 genes in this pathway |
| PID HEDGEHOG 2PATHWAY | 22 | 17 | All SZGR 2.0 genes in this pathway |
| CHEMNITZ RESPONSE TO PROSTAGLANDIN E2 DN | 391 | 222 | All SZGR 2.0 genes in this pathway |
| DAVICIONI TARGETS OF PAX FOXO1 FUSIONS UP | 255 | 177 | All SZGR 2.0 genes in this pathway |
| WIKMAN ASBESTOS LUNG CANCER UP | 17 | 11 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| CHOW RASSF1 TARGETS DN | 29 | 19 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| TERAMOTO OPN TARGETS CLUSTER 4 | 13 | 8 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| FERREIRA EWINGS SARCOMA UNSTABLE VS STABLE DN | 98 | 59 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR UP | 176 | 115 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT AND CANCER BOX4 DN | 32 | 22 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION DN | 517 | 309 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| YAMAZAKI TCEB3 TARGETS UP | 175 | 116 | All SZGR 2.0 genes in this pathway |
| HEDENFALK BREAST CANCER BRCA1 VS BRCA2 | 163 | 113 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 5 | 482 | 296 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER T7 | 98 | 63 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE UP | 578 | 341 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| ALONSO METASTASIS UP | 198 | 128 | All SZGR 2.0 genes in this pathway |
| ROME INSULIN TARGETS IN MUSCLE UP | 442 | 263 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 17 | 181 | 101 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| YANG BCL3 TARGETS UP | 364 | 236 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 | 227 | 149 | All SZGR 2.0 genes in this pathway |