| SCHUETZ BREAST CANCER DUCTAL INVASIVE DN
| 84 | 53 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST DUCTAL CARCINOMA VS DUCTAL NORMAL DN
| 198 | 110 | All SZGR 2.0 genes in this pathway |
| HOOI ST7 TARGETS DN
| 123 | 78 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE UP
| 423 | 283 | All SZGR 2.0 genes in this pathway |
| LIU TARGETS OF VMYB VS CMYB DN
| 43 | 30 | All SZGR 2.0 genes in this pathway |
| LAIHO COLORECTAL CANCER SERRATED UP
| 112 | 71 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY UP
| 430 | 232 | All SZGR 2.0 genes in this pathway |
| JAEGER METASTASIS DN
| 258 | 141 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 6HR DN
| 41 | 29 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 10D UP
| 194 | 122 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 DN
| 242 | 165 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 DN
| 281 | 186 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION SUSTAINED IN GRANULOCYTE UP
| 15 | 9 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC UP
| 185 | 126 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION GRANULOCYTE UP
| 55 | 34 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION MONOCYTE UP
| 204 | 140 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION ERYTHROCYTE UP
| 157 | 104 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION SUSTAINED IN MONOCYTE UP
| 21 | 15 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION SUSTAINDED IN ERYTHROCYTE UP
| 44 | 30 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS UP
| 457 | 269 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP
| 501 | 327 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP
| 557 | 331 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS UP
| 214 | 155 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 8HR UP
| 164 | 122 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 12HR UP
| 162 | 116 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML QUIESCENT VS NORMAL QUIESCENT DN
| 47 | 32 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML DIVIDING VS NORMAL DIVIDING DN
| 10 | 6 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML DIVIDING VS NORMAL QUIESCENT DN
| 95 | 57 | All SZGR 2.0 genes in this pathway |
| GRAHAM NORMAL QUIESCENT VS NORMAL DIVIDING UP
| 66 | 47 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES SKIN UP
| 177 | 113 | All SZGR 2.0 genes in this pathway |
| PROVENZANI METASTASIS UP
| 194 | 112 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 UP
| 329 | 196 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP
| 552 | 347 | All SZGR 2.0 genes in this pathway |
| OUELLET OVARIAN CANCER INVASIVE VS LMP UP
| 117 | 85 | All SZGR 2.0 genes in this pathway |
| APPIERTO RESPONSE TO FENRETINIDE UP
| 38 | 26 | All SZGR 2.0 genes in this pathway |
| SNIJDERS AMPLIFIED IN HEAD AND NECK TUMORS
| 37 | 27 | All SZGR 2.0 genes in this pathway |
| KORKOLA TERATOMA
| 39 | 25 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 DN
| 156 | 106 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 AND HIF1A UP
| 142 | 104 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN
| 1024 | 594 | All SZGR 2.0 genes in this pathway |
| WU CELL MIGRATION
| 184 | 114 | All SZGR 2.0 genes in this pathway |
| GOTZMANN EPITHELIAL TO MESENCHYMAL TRANSITION DN
| 206 | 136 | All SZGR 2.0 genes in this pathway |
| PETRETTO CARDIAC HYPERTROPHY
| 34 | 26 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 240 HELA
| 60 | 43 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 480 HELA
| 164 | 118 | All SZGR 2.0 genes in this pathway |
| AMIT SERUM RESPONSE 60 MCF10A
| 57 | 42 | All SZGR 2.0 genes in this pathway |
| AMIT SERUM RESPONSE 240 MCF10A
| 57 | 36 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO IKK INHIBITOR AND TNF UP
| 223 | 140 | All SZGR 2.0 genes in this pathway |
| KENNY CTNNB1 TARGETS UP
| 50 | 30 | All SZGR 2.0 genes in this pathway |
| ZUCCHI METASTASIS DN
| 44 | 29 | All SZGR 2.0 genes in this pathway |
| SANA TNF SIGNALING DN
| 90 | 57 | All SZGR 2.0 genes in this pathway |
| FRASOR RESPONSE TO ESTRADIOL DN
| 82 | 52 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS
| 882 | 572 | All SZGR 2.0 genes in this pathway |
| BECKER TAMOXIFEN RESISTANCE DN
| 52 | 37 | All SZGR 2.0 genes in this pathway |
| NING CHRONIC OBSTRUCTIVE PULMONARY DISEASE UP
| 157 | 105 | All SZGR 2.0 genes in this pathway |
| LEI MYB TARGETS
| 318 | 215 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN
| 408 | 274 | All SZGR 2.0 genes in this pathway |
| CHEN LVAD SUPPORT OF FAILING HEART DN
| 42 | 32 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS DN
| 371 | 218 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN
| 830 | 547 | All SZGR 2.0 genes in this pathway |
| GAJATE RESPONSE TO TRABECTEDIN UP
| 67 | 45 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN UP
| 262 | 186 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK UP
| 244 | 151 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR UP
| 178 | 111 | All SZGR 2.0 genes in this pathway |
| GENTILE UV HIGH DOSE DN
| 312 | 203 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C1
| 72 | 45 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC UP
| 202 | 115 | All SZGR 2.0 genes in this pathway |
| GENTILE RESPONSE CLUSTER D3
| 61 | 39 | All SZGR 2.0 genes in this pathway |
| RIGGINS TAMOXIFEN RESISTANCE DN
| 220 | 147 | All SZGR 2.0 genes in this pathway |
| CLAUS PGR POSITIVE MENINGIOMA DN
| 12 | 10 | All SZGR 2.0 genes in this pathway |
| YAMASHITA METHYLATED IN PROSTATE CANCER
| 57 | 29 | All SZGR 2.0 genes in this pathway |
| TING SILENCED BY DICER
| 31 | 17 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE DN
| 315 | 197 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN
| 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP
| 648 | 398 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL OPTIMAL DEBULKING
| 246 | 152 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL DN
| 216 | 143 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN
| 546 | 362 | All SZGR 2.0 genes in this pathway |
| HUPER BREAST BASAL VS LUMINAL DN
| 59 | 44 | All SZGR 2.0 genes in this pathway |
| RUIZ TNC TARGETS UP
| 153 | 107 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER ESR1 DN
| 240 | 153 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 36HR UP
| 221 | 150 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR UP
| 293 | 203 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS UP
| 108 | 78 | All SZGR 2.0 genes in this pathway |
| COLINA TARGETS OF 4EBP1 AND 4EBP2
| 356 | 214 | All SZGR 2.0 genes in this pathway |
| KYNG WERNER SYNDROM AND NORMAL AGING DN
| 225 | 124 | All SZGR 2.0 genes in this pathway |
| DAZARD UV RESPONSE CLUSTER G24
| 28 | 19 | All SZGR 2.0 genes in this pathway |
| HIRSCH CELLULAR TRANSFORMATION SIGNATURE UP
| 242 | 159 | All SZGR 2.0 genes in this pathway |
| LU EZH2 TARGETS UP
| 295 | 155 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR DN
| 505 | 328 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS UP
| 259 | 159 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3
| 824 | 528 | All SZGR 2.0 genes in this pathway |
| DELACROIX RARG BOUND MEF
| 367 | 231 | All SZGR 2.0 genes in this pathway |
| DELACROIX RAR TARGETS UP
| 48 | 33 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA VIA KDM3A
| 53 | 34 | All SZGR 2.0 genes in this pathway |
| BOSCO EPITHELIAL DIFFERENTIATION MODULE
| 69 | 31 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF
| 516 | 308 | All SZGR 2.0 genes in this pathway |
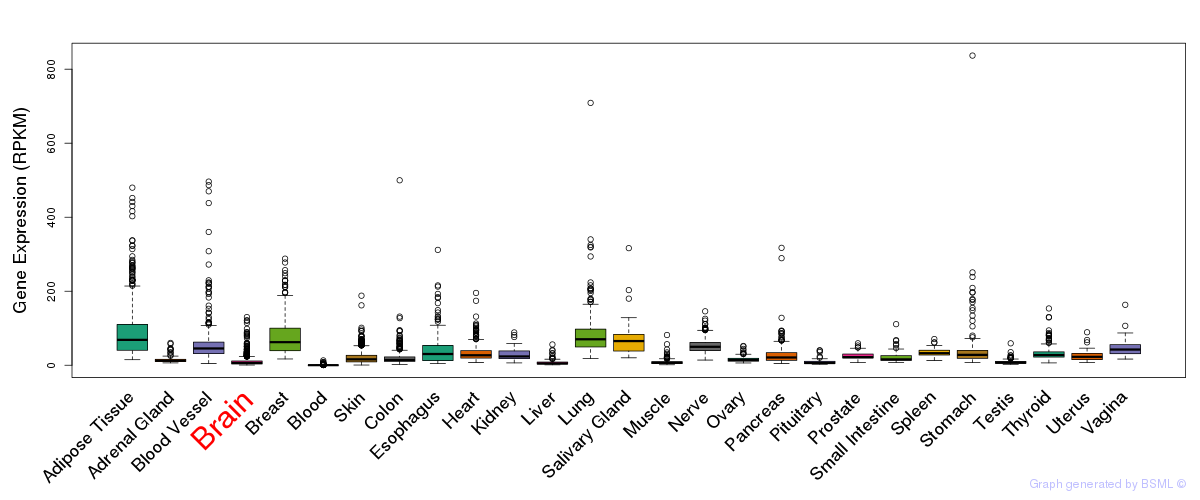
 Differentially methylated gene
Differentially methylated gene eQTL annotation
eQTL annotation