Gene Page: MARCKS
Summary ?
| GeneID | 4082 |
| Symbol | MARCKS |
| Synonyms | 80K-L|MACS|PKCSL|PRKCSL |
| Description | myristoylated alanine rich protein kinase C substrate |
| Reference | MIM:177061|HGNC:HGNC:6759|Ensembl:ENSG00000277443|HPRD:07519|Vega:OTTHUMG00000188327 |
| Gene type | protein-coding |
| Map location | 6q22.2 |
| Pascal p-value | 0.37 |
| Fetal beta | 1.605 |
| eGene | Myers' cis & trans Meta |
| Support | CANABINOID DOPAMINE INTRACELLULAR SIGNAL TRANSDUCTION METABOTROPIC GLUTAMATE RECEPTOR SEROTONIN Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.01 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7835277 | chr8 | 62922905 | MARCKS | 4082 | 0.06 | trans |
Section II. Transcriptome annotation
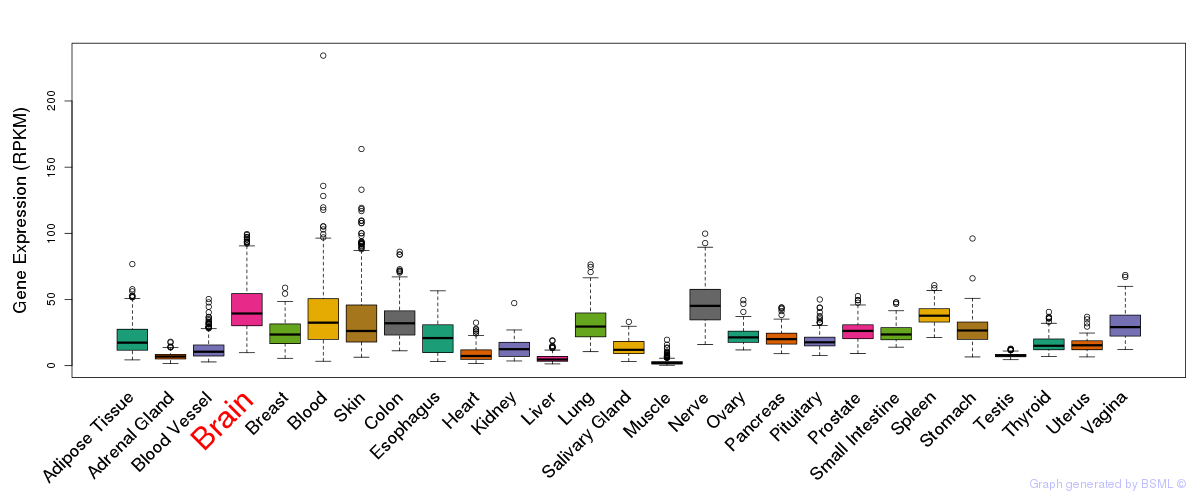
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005516 | calmodulin binding | TAS | 1560845 | |
| GO:0051015 | actin filament binding | TAS | 1560845 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005813 | centrosome | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0016020 | membrane | IEA | - | |
| GO:0005938 | cell cortex | IEA | - | |
| GO:0015629 | actin cytoskeleton | TAS | 1560845 | |
| GO:0042585 | germinal vesicle | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG FC GAMMA R MEDIATED PHAGOCYTOSIS | 97 | 71 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CALCINEURIN PATHWAY | 21 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRATION OF ENERGY METABOLISM | 120 | 84 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF INSULIN SECRETION | 93 | 65 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | 11 | 9 | All SZGR 2.0 genes in this pathway |
| PICCALUGA ANGIOIMMUNOBLASTIC LYMPHOMA UP | 205 | 140 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| DAVICIONI TARGETS OF PAX FOXO1 FUSIONS DN | 68 | 49 | All SZGR 2.0 genes in this pathway |
| DAVICIONI RHABDOMYOSARCOMA PAX FOXO1 FUSION DN | 15 | 11 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS DN | 463 | 290 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE UP | 485 | 293 | All SZGR 2.0 genes in this pathway |
| PUIFFE INVASION INHIBITED BY ASCITES UP | 82 | 51 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| DOANE RESPONSE TO ANDROGEN DN | 241 | 146 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| BILBAN B CLL LPL DN | 42 | 25 | All SZGR 2.0 genes in this pathway |
| HOEBEKE LYMPHOID STEM CELL DN | 86 | 59 | All SZGR 2.0 genes in this pathway |
| DITTMER PTHLH TARGETS UP | 112 | 68 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA UP | 544 | 308 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LPS UP | 431 | 237 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION DN | 329 | 219 | All SZGR 2.0 genes in this pathway |
| BIDUS METASTASIS UP | 214 | 134 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES SKIN UP | 177 | 113 | All SZGR 2.0 genes in this pathway |
| LANDIS ERBB2 BREAST TUMORS 65 DN | 37 | 22 | All SZGR 2.0 genes in this pathway |
| CONCANNON APOPTOSIS BY EPOXOMICIN DN | 172 | 112 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| LANDIS ERBB2 BREAST TUMORS 324 DN | 149 | 93 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| JOHANSSON GLIOMAGENESIS BY PDGFB UP | 58 | 44 | All SZGR 2.0 genes in this pathway |
| IVANOV MUTATED IN COLON CANCER | 13 | 9 | All SZGR 2.0 genes in this pathway |
| LI CISPLATIN RESISTANCE DN | 35 | 20 | All SZGR 2.0 genes in this pathway |
| HERNANDEZ ABERRANT MITOSIS BY DOCETACEL 2NM UP | 81 | 57 | All SZGR 2.0 genes in this pathway |
| DIRMEIER LMP1 RESPONSE LATE UP | 57 | 41 | All SZGR 2.0 genes in this pathway |
| YANAGIHARA ESX1 TARGETS | 30 | 19 | All SZGR 2.0 genes in this pathway |
| WEINMANN ADAPTATION TO HYPOXIA UP | 29 | 24 | All SZGR 2.0 genes in this pathway |
| WEINMANN ADAPTATION TO HYPOXIA DN | 41 | 33 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS DN | 193 | 112 | All SZGR 2.0 genes in this pathway |
| SIMBULAN UV RESPONSE NORMAL DN | 33 | 27 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 UP | 209 | 139 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 ONLY DN | 44 | 30 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| FRIDMAN SENESCENCE DN | 13 | 6 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION UP | 195 | 138 | All SZGR 2.0 genes in this pathway |
| MORI SMALL PRE BII LYMPHOCYTE UP | 86 | 57 | All SZGR 2.0 genes in this pathway |
| MORI MATURE B LYMPHOCYTE DN | 75 | 43 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 UP | 256 | 159 | All SZGR 2.0 genes in this pathway |
| BROCKE APOPTOSIS REVERSED BY IL6 | 144 | 98 | All SZGR 2.0 genes in this pathway |
| LE EGR2 TARGETS UP | 108 | 75 | All SZGR 2.0 genes in this pathway |
| OKUMURA INFLAMMATORY RESPONSE LPS | 183 | 115 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA CD2 UP | 45 | 32 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED UP | 183 | 111 | All SZGR 2.0 genes in this pathway |
| NGUYEN NOTCH1 TARGETS DN | 86 | 67 | All SZGR 2.0 genes in this pathway |
| KUMAR TARGETS OF MLL AF9 FUSION | 405 | 264 | All SZGR 2.0 genes in this pathway |
| JIANG AGING CEREBRAL CORTEX UP | 36 | 27 | All SZGR 2.0 genes in this pathway |
| CHEN LVAD SUPPORT OF FAILING HEART DN | 42 | 32 | All SZGR 2.0 genes in this pathway |
| PAL PRMT5 TARGETS UP | 203 | 135 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR DN | 298 | 200 | All SZGR 2.0 genes in this pathway |
| ZHU CMV 8 HR DN | 53 | 40 | All SZGR 2.0 genes in this pathway |
| TAKAO RESPONSE TO UVB RADIATION DN | 98 | 67 | All SZGR 2.0 genes in this pathway |
| ZHU CMV ALL DN | 128 | 93 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS PEAK AT 0HR | 63 | 48 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 11 | 57 | 40 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA NORMAL | 311 | 205 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D8 | 40 | 29 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS UP | 280 | 183 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE DN | 373 | 196 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER T7 | 98 | 63 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS DN | 105 | 63 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN | 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN DN | 353 | 226 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR UP | 174 | 96 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN UP | 439 | 257 | All SZGR 2.0 genes in this pathway |
| CHEN HOXA5 TARGETS 9HR DN | 41 | 17 | All SZGR 2.0 genes in this pathway |
| QI PLASMACYTOMA UP | 259 | 185 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP | 374 | 247 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER SURVIVAL DN | 175 | 103 | All SZGR 2.0 genes in this pathway |
| DAVIES MULTIPLE MYELOMA VS MGUS DN | 28 | 18 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA D DN | 78 | 34 | All SZGR 2.0 genes in this pathway |
| CHAUHAN RESPONSE TO METHOXYESTRADIOL DN | 102 | 65 | All SZGR 2.0 genes in this pathway |
| LINDSTEDT DENDRITIC CELL MATURATION B | 53 | 36 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 UP | 338 | 225 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF UP | 418 | 282 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 DN | 245 | 150 | All SZGR 2.0 genes in this pathway |
| VANASSE BCL2 TARGETS DN | 74 | 50 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| HOFFMANN IMMATURE TO MATURE B LYMPHOCYTE DN | 50 | 36 | All SZGR 2.0 genes in this pathway |
| LEE RECENT THYMIC EMIGRANT | 227 | 128 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| ZHAN V1 LATE DIFFERENTIATION GENES UP | 32 | 25 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G3 UP | 188 | 121 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS PROLIFERATION UP | 178 | 108 | All SZGR 2.0 genes in this pathway |
| WOO LIVER CANCER RECURRENCE UP | 105 | 75 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS | 535 | 325 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA PCA3 DN | 69 | 38 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA UP | 207 | 143 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 9 | 76 | 45 | All SZGR 2.0 genes in this pathway |
| BAE BRCA1 TARGETS UP | 75 | 47 | All SZGR 2.0 genes in this pathway |
| LI INDUCED T TO NATURAL KILLER DN | 116 | 83 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA PRONEURAL | 177 | 132 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS GROWING | 243 | 155 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR DN | 505 | 328 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS DN | 229 | 135 | All SZGR 2.0 genes in this pathway |
| PASINI SUZ12 TARGETS DN | 315 | 215 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| IKEDA MIR30 TARGETS UP | 116 | 87 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 10HR DN | 244 | 157 | All SZGR 2.0 genes in this pathway |
| ALFANO MYC TARGETS | 239 | 156 | All SZGR 2.0 genes in this pathway |
| HOLLEMAN ASPARAGINASE RESISTANCE ALL DN | 25 | 14 | All SZGR 2.0 genes in this pathway |
| DURAND STROMA S UP | 297 | 194 | All SZGR 2.0 genes in this pathway |
| ZWANG EGF PERSISTENTLY DN | 61 | 36 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-128 | 801 | 807 | 1A | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-138 | 783 | 789 | 1A | hsa-miR-138brain | AGCUGGUGUUGUGAAUC |
| miR-142-3p | 555 | 562 | 1A,m8 | hsa-miR-142-3p | UGUAGUGUUUCCUACUUUAUGGA |
| miR-143 | 191 | 197 | 1A | hsa-miR-143brain | UGAGAUGAAGCACUGUAGCUCA |
| miR-144 | 961 | 967 | m8 | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| miR-181 | 1124 | 1130 | 1A | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-182 | 455 | 462 | 1A,m8 | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| miR-200bc/429 | 293 | 300 | 1A,m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-205 | 373 | 380 | 1A,m8 | hsa-miR-205 | UCCUUCAUUCCACCGGAGUCUG |
| miR-218 | 839 | 845 | m8 | hsa-miR-218brain | UUGUGCUUGAUCUAACCAUGU |
| miR-23 | 1210 | 1217 | 1A,m8 | hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC |
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| miR-27 | 801 | 807 | m8 | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-299-5p | 655 | 661 | 1A | hsa-miR-299-5p | UGGUUUACCGUCCCACAUACAU |
| miR-30-5p | 548 | 554 | 1A | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-323 | 1210 | 1216 | 1A | hsa-miR-323brain | GCACAUUACACGGUCGACCUCU |
| miR-370 | 785 | 791 | m8 | hsa-miR-370brain | GCCUGCUGGGGUGGAACCUGG |
| miR-381 | 811 | 817 | 1A | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU | ||||
| miR-421 | 540 | 546 | 1A | hsa-miR-421 | GGCCUCAUUAAAUGUUUGUUG |
| miR-491 | 178 | 185 | 1A,m8 | hsa-miR-491brain | AGUGGGGAACCCUUCCAUGAGGA |
| miR-495 | 44 | 50 | m8 | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
| hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU | ||||
| miR-499 | 638 | 644 | m8 | hsa-miR-499 | UUAAGACUUGCAGUGAUGUUUAA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.