Gene Page: CXCL9
Summary ?
| GeneID | 4283 |
| Symbol | CXCL9 |
| Synonyms | CMK|Humig|MIG|SCYB9|crg-10 |
| Description | C-X-C motif chemokine ligand 9 |
| Reference | MIM:601704|HGNC:HGNC:7098|Ensembl:ENSG00000138755|HPRD:03416|Vega:OTTHUMG00000160889 |
| Gene type | protein-coding |
| Map location | 4q21 |
| Pascal p-value | 0.071 |
| Fetal beta | -0.327 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs12041093 | chr1 | 58462509 | CXCL9 | 4283 | 0.02 | trans | ||
| rs12023109 | chr1 | 58482942 | CXCL9 | 4283 | 0.02 | trans | ||
| rs1342707 | chr1 | 147264296 | CXCL9 | 4283 | 1.103E-5 | trans | ||
| rs3766034 | chr1 | 169087664 | CXCL9 | 4283 | 0.12 | trans | ||
| rs17013505 | chr1 | 208999490 | CXCL9 | 4283 | 0.17 | trans | ||
| rs6656512 | chr1 | 223644438 | CXCL9 | 4283 | 0.2 | trans | ||
| rs1342362 | chr1 | 242551697 | CXCL9 | 4283 | 0.1 | trans | ||
| rs17042744 | chr2 | 21824821 | CXCL9 | 4283 | 0.01 | trans | ||
| rs918897 | chr2 | 21825859 | CXCL9 | 4283 | 0.17 | trans | ||
| rs2113625 | chr2 | 21828617 | CXCL9 | 4283 | 0.01 | trans | ||
| rs17680142 | chr2 | 21849670 | CXCL9 | 4283 | 0.01 | trans | ||
| rs6547745 | chr2 | 21852551 | CXCL9 | 4283 | 0.01 | trans | ||
| rs17042868 | chr2 | 21895688 | CXCL9 | 4283 | 0.01 | trans | ||
| rs11893111 | chr2 | 21910980 | CXCL9 | 4283 | 0.01 | trans | ||
| rs529098 | chr3 | 54677190 | CXCL9 | 4283 | 0.05 | trans | ||
| rs510605 | chr3 | 54709161 | CXCL9 | 4283 | 0.06 | trans | ||
| rs666071 | chr3 | 54709720 | CXCL9 | 4283 | 0.13 | trans | ||
| rs17576136 | chr4 | 37384794 | CXCL9 | 4283 | 0.05 | trans | ||
| rs4516856 | chr5 | 43889206 | CXCL9 | 4283 | 0.13 | trans | ||
| rs16869716 | chr6 | 71743471 | CXCL9 | 4283 | 1.342E-7 | trans | ||
| rs7464123 | chr8 | 66491542 | CXCL9 | 4283 | 0.09 | trans | ||
| rs16932340 | chr8 | 66684243 | CXCL9 | 4283 | 0.01 | trans | ||
| rs7861200 | chr9 | 1833109 | CXCL9 | 4283 | 0.18 | trans | ||
| rs10960702 | chr9 | 12540168 | CXCL9 | 4283 | 0.01 | trans | ||
| rs11829144 | chr12 | 10350032 | CXCL9 | 4283 | 0.2 | trans | ||
| rs11836020 | chr12 | 67610912 | CXCL9 | 4283 | 0.18 | trans | ||
| rs11613629 | chr12 | 104972976 | CXCL9 | 4283 | 0 | trans | ||
| rs10484008 | chr14 | 89376562 | CXCL9 | 4283 | 2.951E-4 | trans | ||
| rs4303487 | chr16 | 90033254 | CXCL9 | 4283 | 0.16 | trans | ||
| rs17701187 | chr17 | 15256066 | CXCL9 | 4283 | 0.17 | trans | ||
| rs8098388 | chr18 | 24080746 | CXCL9 | 4283 | 0.01 | trans | ||
| rs16966813 | chr19 | 32698878 | CXCL9 | 4283 | 0.09 | trans | ||
| rs4802583 | chr19 | 39025340 | CXCL9 | 4283 | 0.05 | trans | ||
| rs17819434 | chr22 | 20922469 | CXCL9 | 4283 | 0.18 | trans | ||
| rs9619990 | chr22 | 42163258 | CXCL9 | 4283 | 0.01 | trans | ||
| rs8141783 | chr22 | 42179110 | CXCL9 | 4283 | 0.01 | trans | ||
| rs16995164 | chrX | 148536507 | CXCL9 | 4283 | 0.18 | trans |
Section II. Transcriptome annotation
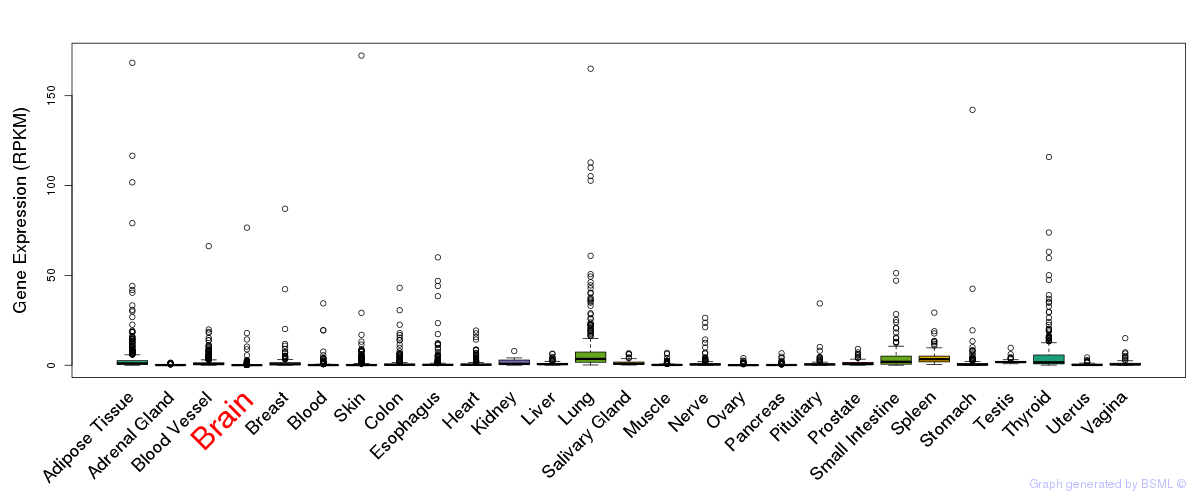
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CYTOKINE CYTOKINE RECEPTOR INTERACTION | 267 | 161 | All SZGR 2.0 genes in this pathway |
| KEGG CHEMOKINE SIGNALING PATHWAY | 190 | 128 | All SZGR 2.0 genes in this pathway |
| KEGG TOLL LIKE RECEPTOR SIGNALING PATHWAY | 102 | 88 | All SZGR 2.0 genes in this pathway |
| PID IL23 PATHWAY | 37 | 30 | All SZGR 2.0 genes in this pathway |
| PID CXCR3 PATHWAY | 43 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME PEPTIDE LIGAND BINDING RECEPTORS | 188 | 108 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | 305 | 177 | All SZGR 2.0 genes in this pathway |
| REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | 57 | 27 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA I SIGNALLING EVENTS | 195 | 114 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR LIGAND BINDING | 408 | 246 | All SZGR 2.0 genes in this pathway |
| PICCALUGA ANGIOIMMUNOBLASTIC LYMPHOMA UP | 205 | 140 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA UP | 294 | 178 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS DN | 463 | 290 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LPS UP | 431 | 237 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| KOINUMA COLON CANCER MSI UP | 16 | 11 | All SZGR 2.0 genes in this pathway |
| WONG ENDOMETRIAL CANCER LATE | 10 | 5 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 6 | 84 | 54 | All SZGR 2.0 genes in this pathway |
| WATTEL AUTONOMOUS THYROID ADENOMA DN | 55 | 29 | All SZGR 2.0 genes in this pathway |
| MANTOVANI NFKB TARGETS UP | 43 | 33 | All SZGR 2.0 genes in this pathway |
| MANTOVANI VIRAL GPCR SIGNALING UP | 86 | 54 | All SZGR 2.0 genes in this pathway |
| SHIN B CELL LYMPHOMA CLUSTER 1 | 13 | 11 | All SZGR 2.0 genes in this pathway |
| WIELAND UP BY HBV INFECTION | 101 | 66 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC E2F1 UP | 56 | 34 | All SZGR 2.0 genes in this pathway |
| SANA RESPONSE TO IFNG UP | 78 | 50 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| NATSUME RESPONSE TO INTERFERON BETA UP | 71 | 49 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 16HR DN | 86 | 62 | All SZGR 2.0 genes in this pathway |
| RHODES UNDIFFERENTIATED CANCER | 69 | 44 | All SZGR 2.0 genes in this pathway |
| MCLACHLAN DENTAL CARIES DN | 245 | 144 | All SZGR 2.0 genes in this pathway |
| NIELSEN LEIOMYOSARCOMA UP | 18 | 10 | All SZGR 2.0 genes in this pathway |
| MCLACHLAN DENTAL CARIES UP | 253 | 147 | All SZGR 2.0 genes in this pathway |
| KIM LRRC3B TARGETS | 30 | 24 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE UP | 156 | 92 | All SZGR 2.0 genes in this pathway |
| WALLACE PROSTATE CANCER RACE UP | 299 | 167 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BRAIN UP | 39 | 20 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN PLEURA DN | 27 | 15 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| GAURNIER PSMD4 TARGETS | 73 | 55 | All SZGR 2.0 genes in this pathway |
| AUJLA IL22 AND IL17A SIGNALING | 11 | 5 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS UP | 165 | 118 | All SZGR 2.0 genes in this pathway |
| ICHIBA GRAFT VERSUS HOST DISEASE D7 UP | 107 | 67 | All SZGR 2.0 genes in this pathway |
| ICHIBA GRAFT VERSUS HOST DISEASE 35D UP | 131 | 79 | All SZGR 2.0 genes in this pathway |
| WUNDER INFLAMMATORY RESPONSE AND CHOLESTEROL DN | 12 | 7 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 UP | 338 | 225 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF UP | 418 | 282 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| POOLA INVASIVE BREAST CANCER UP | 288 | 168 | All SZGR 2.0 genes in this pathway |
| KUROZUMI RESPONSE TO ONCOCYTIC VIRUS | 44 | 30 | All SZGR 2.0 genes in this pathway |
| KUROZUMI RESPONSE TO ONCOCYTIC VIRUS AND CYCLIC RGD | 21 | 15 | All SZGR 2.0 genes in this pathway |
| WORSCHECH TUMOR EVASION AND TOLEROGENICITY UP | 30 | 19 | All SZGR 2.0 genes in this pathway |
| NAKAYAMA SOFT TISSUE TUMORS PCA1 UP | 76 | 46 | All SZGR 2.0 genes in this pathway |
| LI INDUCED T TO NATURAL KILLER UP | 307 | 182 | All SZGR 2.0 genes in this pathway |
| SERVITJA LIVER HNF1A TARGETS DN | 157 | 105 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
| BOSCO TH1 CYTOTOXIC MODULE | 114 | 62 | All SZGR 2.0 genes in this pathway |
| NABA SECRETED FACTORS | 344 | 197 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |