Gene Page: NDN
Summary ?
| GeneID | 4692 |
| Symbol | NDN |
| Synonyms | HsT16328|PWCR |
| Description | necdin, MAGE family member |
| Reference | MIM:602117|HGNC:HGNC:7675|Ensembl:ENSG00000182636|HPRD:03667|Vega:OTTHUMG00000129161 |
| Gene type | protein-coding |
| Map location | 15q11.2 |
| Pascal p-value | 0.648 |
| Sherlock p-value | 0.65 |
| Fetal beta | -0.054 |
| eGene | Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CNV:YES | Copy number variation studies | Manual curation | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 6 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
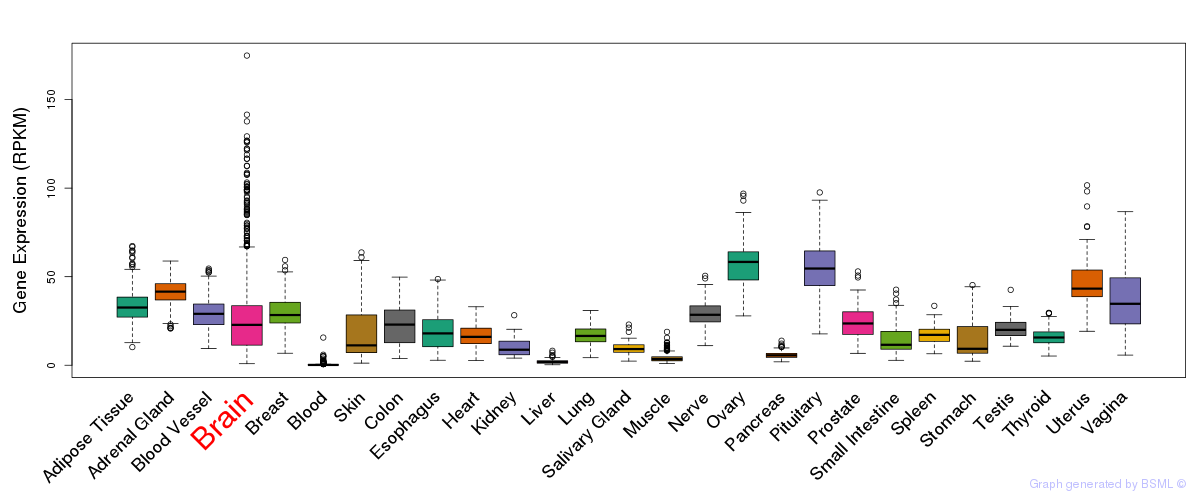
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NHP2 | 0.87 | 0.85 |
| MRPS24 | 0.84 | 0.79 |
| C12orf10 | 0.83 | 0.80 |
| AC174470.1 | 0.83 | 0.79 |
| GADD45GIP1 | 0.83 | 0.80 |
| TOMM5 | 0.83 | 0.81 |
| RABAC1 | 0.82 | 0.78 |
| POP7 | 0.82 | 0.79 |
| EXOSC4 | 0.82 | 0.81 |
| CCDC56 | 0.82 | 0.79 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.26 | -0.61 | -0.61 |
| AF347015.2 | -0.58 | -0.57 |
| AF347015.15 | -0.57 | -0.58 |
| AF347015.33 | -0.57 | -0.58 |
| MT-CYB | -0.56 | -0.57 |
| AF347015.8 | -0.56 | -0.56 |
| MT-CO2 | -0.52 | -0.51 |
| AF347015.18 | -0.51 | -0.54 |
| AF347015.9 | -0.51 | -0.56 |
| FAM38A | -0.51 | -0.52 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003677 | DNA binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 17353931 | |
| GO:0043015 | gamma-tubulin binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007417 | central nervous system development | IEA | Brain (GO term level: 6) | - |
| GO:0007413 | axonal fasciculation | IEA | neuron, axon (GO term level: 13) | - |
| GO:0001764 | neuron migration | IEA | neuron (GO term level: 8) | - |
| GO:0048676 | axon extension involved in development | IEA | axon (GO term level: 14) | - |
| GO:0048666 | neuron development | IEA | neuron (GO term level: 9) | - |
| GO:0008347 | glial cell migration | IEA | neuron, Glial (GO term level: 8) | - |
| GO:0001558 | regulation of cell growth | IEA | - | |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| GO:0008285 | negative regulation of cell proliferation | TAS | 9630521 | |
| GO:0007585 | respiratory gaseous exchange | IEA | - | |
| GO:0048011 | nerve growth factor receptor signaling pathway | IEA | - | |
| GO:0019233 | sensory perception of pain | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005813 | centrosome | IEA | - | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0043204 | perikaryon | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ARSE | CDPX | CDPX1 | CDPXR | MGC163310 | arylsulfatase E (chondrodysplasia punctata 1) | Two-hybrid | BioGRID | 16169070 |
| AUP1 | - | ancient ubiquitous protein 1 | Affinity Capture-MS | BioGRID | 17353931 |
| DHCR7 | SLOS | 7-dehydrocholesterol reductase | Affinity Capture-MS | BioGRID | 17353931 |
| E2F1 | E2F-1 | RBAP1 | RBBP3 | RBP3 | E2F transcription factor 1 | - | HPRD,BioGRID | 9422723 |14593116 |
| E2F4 | E2F-4 | E2F transcription factor 4, p107/p130-binding | - | HPRD | 12198120 |
| HNRNPU | HNRPU | SAF-A | U21.1 | heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) | - | HPRD,BioGRID | 11813259 |
| IL1A | IL-1A | IL1 | IL1-ALPHA | IL1F1 | interleukin 1, alpha | - | HPRD,BioGRID | 12913118 |
| LASS2 | CerS2 | FLJ10243 | L3 | MGC987 | SP260 | TMSG1 | LAG1 homolog, ceramide synthase 2 | Affinity Capture-MS | BioGRID | 17353931 |
| NDN | HsT16328 | PWCR | necdin homolog (mouse) | - | HPRD | 12414813 |
| NDN | HsT16328 | PWCR | necdin homolog (mouse) | Two-hybrid | BioGRID | 10915798 |
| NGFR | CD271 | Gp80-LNGFR | TNFRSF16 | p75(NTR) | p75NTR | nerve growth factor receptor (TNFR superfamily, member 16) | - | HPRD,BioGRID | 12414813 |12716928 |14593116 |
| NUBP1 | MGC117406 | MGC130052 | MGC130053 | NBP | NBP1 | nucleotide binding protein 1 (MinD homolog, E. coli) | - | HPRD,BioGRID | 10915798 |
| NUCB2 | NEFA | nucleobindin 2 | - | HPRD,BioGRID | 10915798 |
| PJA1 | RNF70 | praja 1 | - | HPRD,BioGRID | 11959851 |
| RPS11 | - | ribosomal protein S11 | Affinity Capture-MS | BioGRID | 17353931 |
| SEC61A1 | HSEC61 | SEC61 | SEC61A | Sec61 alpha 1 subunit (S. cerevisiae) | Affinity Capture-MS | BioGRID | 17353931 |
| SRPRB | APMCF1 | signal recognition particle receptor, B subunit | Affinity Capture-MS | BioGRID | 17353931 |
| TMEM33 | 1600019D15Rik | FLJ10525 | SHINC3 | transmembrane protein 33 | Affinity Capture-MS | BioGRID | 17353931 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | - | HPRD,BioGRID | 10347180 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID P75 NTR PATHWAY | 69 | 51 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| SCHUETZ BREAST CANCER DUCTAL INVASIVE UP | 351 | 230 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| HOEBEKE LYMPHOID STEM CELL DN | 86 | 59 | All SZGR 2.0 genes in this pathway |
| SABATES COLORECTAL ADENOMA DN | 291 | 176 | All SZGR 2.0 genes in this pathway |
| JAATINEN HEMATOPOIETIC STEM CELL UP | 316 | 190 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER LMP DN | 199 | 124 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS G UP | 238 | 135 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 4 5WK DN | 196 | 131 | All SZGR 2.0 genes in this pathway |
| WONG ENDMETRIUM CANCER DN | 82 | 53 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION APOBEC2 | 27 | 19 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION 12HR | 43 | 35 | All SZGR 2.0 genes in this pathway |
| JOHANSSON BRAIN CANCER EARLY VS LATE DN | 45 | 35 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM4 | 261 | 153 | All SZGR 2.0 genes in this pathway |
| GARCIA TARGETS OF FLI1 AND DAX1 DN | 176 | 104 | All SZGR 2.0 genes in this pathway |
| FRIDMAN SENESCENCE UP | 77 | 60 | All SZGR 2.0 genes in this pathway |
| FRIDMAN IMMORTALIZATION DN | 34 | 24 | All SZGR 2.0 genes in this pathway |
| KANNAN TP53 TARGETS UP | 58 | 40 | All SZGR 2.0 genes in this pathway |
| PEART HDAC PROLIFERATION CLUSTER UP | 57 | 35 | All SZGR 2.0 genes in this pathway |
| POMEROY MEDULLOBLASTOMA PROGNOSIS UP | 47 | 30 | All SZGR 2.0 genes in this pathway |
| GUO HEX TARGETS UP | 81 | 54 | All SZGR 2.0 genes in this pathway |
| AFFAR YY1 TARGETS DN | 234 | 137 | All SZGR 2.0 genes in this pathway |
| PAL PRMT5 TARGETS UP | 203 | 135 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION MUSCLE UP | 43 | 33 | All SZGR 2.0 genes in this pathway |
| LEE AGING MUSCLE DN | 46 | 29 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| TSENG IRS1 TARGETS UP | 113 | 71 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| KRASNOSELSKAYA ILF3 TARGETS DN | 46 | 38 | All SZGR 2.0 genes in this pathway |
| TSENG ADIPOGENIC POTENTIAL UP | 30 | 19 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN DN | 353 | 226 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| IZADPANAH STEM CELL ADIPOSE VS BONE DN | 108 | 68 | All SZGR 2.0 genes in this pathway |
| SCHRAETS MLL TARGETS UP | 35 | 21 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| LEE INTRATHYMIC T PROGENITOR | 21 | 14 | All SZGR 2.0 genes in this pathway |
| COLINA TARGETS OF 4EBP1 AND 4EBP2 | 356 | 214 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA UP | 207 | 143 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT UP | 166 | 105 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| NAKAYAMA SOFT TISSUE TUMORS PCA1 DN | 74 | 47 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |
| BRIDEAU IMPRINTED GENES | 63 | 47 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-200bc/429 | 92 | 98 | m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-217 | 90 | 96 | 1A | hsa-miR-217 | UACUGCAUCAGGAACUGAUUGGAU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.