Gene Page: ATP1A3
Summary ?
| GeneID | 478 |
| Symbol | ATP1A3 |
| Synonyms | AHC2|CAPOS|DYT12|RDP |
| Description | ATPase Na+/K+ transporting subunit alpha 3 |
| Reference | MIM:182350|HGNC:HGNC:801|Ensembl:ENSG00000105409|HPRD:01666|Vega:OTTHUMG00000137384 |
| Gene type | protein-coding |
| Map location | 19q13.31 |
| Pascal p-value | 0.195 |
| Sherlock p-value | 0.461 |
| Fetal beta | -1.301 |
| DMG | 1 (# studies) |
| Support | ION BALANCE G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.human_clathrin G2Cdb.human_mGluR5 G2Cdb.human_Synaptosome G2Cdb.humanNRC G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg10657367 | 19 | 42497766 | ATP1A3 | 1.268E-4 | 0.524 | 0.03 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
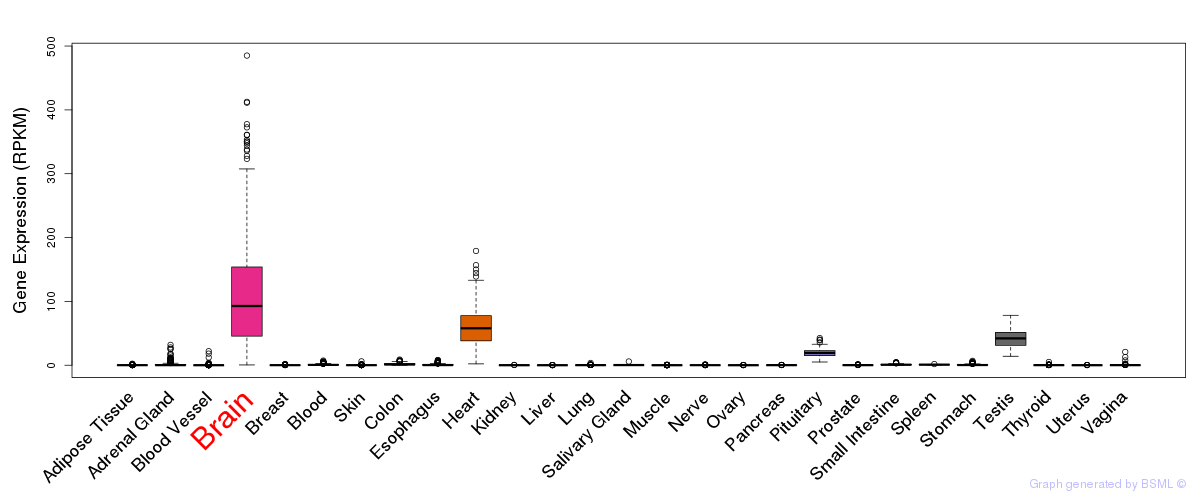
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| IDH3A | 0.92 | 0.91 |
| AC005277.1 | 0.91 | 0.91 |
| NAP1L2 | 0.90 | 0.94 |
| MOAP1 | 0.90 | 0.91 |
| NSF | 0.90 | 0.90 |
| MAP1A | 0.90 | 0.89 |
| MAPK9 | 0.90 | 0.92 |
| NDRG3 | 0.90 | 0.88 |
| SNX10 | 0.89 | 0.92 |
| CNTNAP1 | 0.89 | 0.90 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| BCL7C | -0.55 | -0.60 |
| NME4 | -0.52 | -0.60 |
| RPL28 | -0.48 | -0.54 |
| RPS19P3 | -0.48 | -0.53 |
| HYAL2 | -0.48 | -0.46 |
| RPL23A | -0.47 | -0.47 |
| SH3BP2 | -0.47 | -0.46 |
| RPL18 | -0.47 | -0.50 |
| RPS16P1 | -0.46 | -0.53 |
| RPL37 | -0.46 | -0.56 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0000287 | magnesium ion binding | IEA | - | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0005524 | ATP binding | NAS | 15260953 | |
| GO:0005391 | sodium:potassium-exchanging ATPase activity | IMP | 15260953 | |
| GO:0016787 | hydrolase activity | IEA | - | |
| GO:0016820 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances | IEA | - | |
| GO:0015662 | ATPase activity, coupled to transmembrane movement of ions, phosphorylative mechanism | IEA | - | |
| GO:0015077 | monovalent inorganic cation transmembrane transporter activity | IEA | - | |
| GO:0030955 | potassium ion binding | IEA | - | |
| GO:0031402 | sodium ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0035235 | ionotropic glutamate receptor signaling pathway | IEA | glutamate (GO term level: 8) | - |
| GO:0008344 | adult locomotory behavior | IEA | - | |
| GO:0008542 | visual learning | IEA | - | |
| GO:0008152 | metabolic process | IEA | - | |
| GO:0007613 | memory | IEA | - | |
| GO:0006814 | sodium ion transport | IEA | - | |
| GO:0006813 | potassium ion transport | IEA | - | |
| GO:0042493 | response to drug | IEA | - | |
| GO:0030317 | sperm motility | ISS | - | |
| GO:0030641 | regulation of cellular pH | ISS | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005794 | Golgi apparatus | IDA | 15260953 | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005783 | endoplasmic reticulum | IDA | 15260953 | |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0016021 | integral to membrane | NAS | 15260953 | |
| GO:0005886 | plasma membrane | IDA | 15260953 | |
| GO:0005890 | sodium:potassium-exchanging ATPase complex | IC | 15260953 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CARDIAC MUSCLE CONTRACTION | 80 | 51 | All SZGR 2.0 genes in this pathway |
| KEGG ALDOSTERONE REGULATED SODIUM REABSORPTION | 42 | 29 | All SZGR 2.0 genes in this pathway |
| KEGG PROXIMAL TUBULE BICARBONATE RECLAMATION | 23 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMEMBRANE TRANSPORT OF SMALL MOLECULES | 413 | 270 | All SZGR 2.0 genes in this pathway |
| REACTOME ION TRANSPORT BY P TYPE ATPASES | 34 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME ION CHANNEL TRANSPORT | 55 | 42 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 3 4WK UP | 214 | 144 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 4 5WK UP | 271 | 175 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 5 6WK DN | 137 | 97 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 6 7WK UP | 197 | 135 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION UP | 195 | 138 | All SZGR 2.0 genes in this pathway |
| SMITH TERT TARGETS DN | 87 | 69 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR UP | 240 | 152 | All SZGR 2.0 genes in this pathway |
| CHEN LVAD SUPPORT OF FAILING HEART DN | 42 | 32 | All SZGR 2.0 genes in this pathway |
| KUNINGER IGF1 VS PDGFB TARGETS DN | 46 | 28 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN WITHOUT MGMT 48HR DN | 32 | 25 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN MGMT 48HR DN | 161 | 105 | All SZGR 2.0 genes in this pathway |
| SARRIO EPITHELIAL MESENCHYMAL TRANSITION DN | 154 | 101 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 | 491 | 319 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA PRONEURAL | 177 | 132 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-185 | 168 | 174 | m8 | hsa-miR-185brain | UGGAGAGAAAGGCAGUUC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.