Gene Page: NPTX1
Summary ?
| GeneID | 4884 |
| Symbol | NPTX1 |
| Synonyms | NP1 |
| Description | neuronal pentraxin 1 |
| Reference | MIM:602367|HGNC:HGNC:7952|Ensembl:ENSG00000171246|HPRD:03843|Vega:OTTHUMG00000177607 |
| Gene type | protein-coding |
| Map location | 17q25.3 |
| Pascal p-value | 4.757E-5 |
| Sherlock p-value | 0.911 |
| DMG | 1 (# studies) |
| eGene | Cerebellum Nucleus accumbens basal ganglia Myers' cis & trans |
| Support | INTRACELLULAR SIGNAL TRANSDUCTION G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_clathrin G2Cdb.human_Synaptosome Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg23752000 | 17 | 78442298 | NPTX1 | 1.83E-5 | -0.573 | 0.016 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs8081537 | chr17 | 77708263 | NPTX1 | 4884 | 0.12 | cis | ||
| rs9515552 | chr13 | 89948963 | NPTX1 | 4884 | 0.14 | trans | ||
| rs7321205 | chr13 | 89949815 | NPTX1 | 4884 | 0.09 | trans |
Section II. Transcriptome annotation
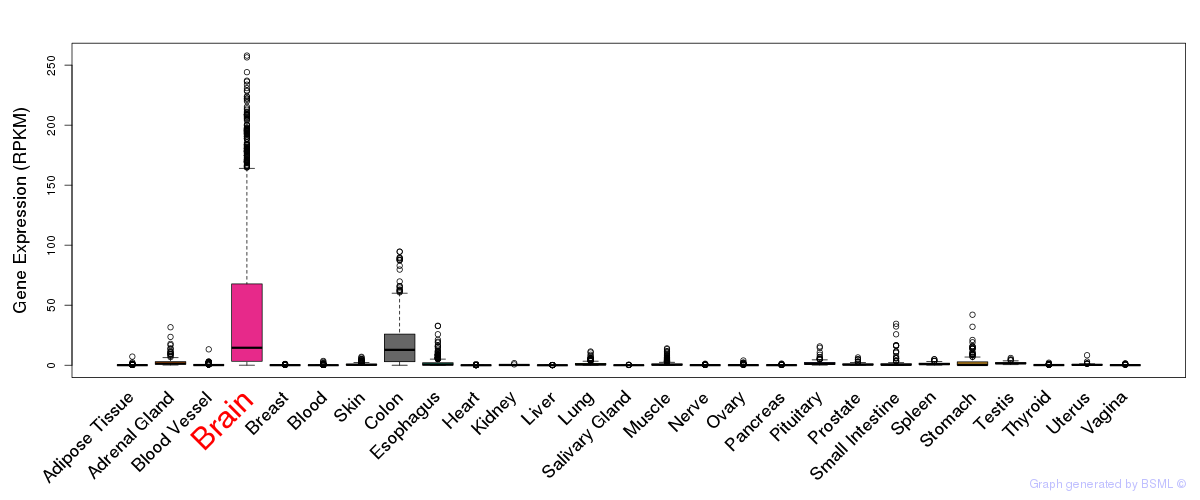
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ITGAV | 0.82 | 0.82 |
| ARHGAP5 | 0.80 | 0.78 |
| GPM6B | 0.79 | 0.76 |
| IL6ST | 0.79 | 0.77 |
| KIAA0494 | 0.79 | 0.78 |
| NEBL | 0.78 | 0.75 |
| ALDH5A1 | 0.77 | 0.77 |
| PCGF5 | 0.76 | 0.72 |
| SLC30A1 | 0.76 | 0.76 |
| SPRED1 | 0.76 | 0.77 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RP9P | -0.41 | -0.51 |
| SNHG12 | -0.41 | -0.48 |
| ST20 | -0.39 | -0.43 |
| MRPL52 | -0.37 | -0.38 |
| BCL7C | -0.37 | -0.42 |
| RPL35 | -0.36 | -0.44 |
| IL32 | -0.35 | -0.35 |
| AL022328.1 | -0.35 | -0.36 |
| RPL36 | -0.35 | -0.43 |
| AC008763.1 | -0.35 | -0.36 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005509 | calcium ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007417 | central nervous system development | TAS | Brain (GO term level: 6) | 8884281 |
| GO:0007268 | synaptic transmission | TAS | neuron, Synap, Neurotransmitter (GO term level: 6) | 8884281 |
| GO:0006810 | transport | TAS | 8884281 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0031410 | cytoplasmic vesicle | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| ZHONG RESPONSE TO AZACITIDINE AND TSA UP | 183 | 119 | All SZGR 2.0 genes in this pathway |
| HUTTMANN B CLL POOR SURVIVAL UP | 276 | 187 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC UP | 185 | 126 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS UP | 149 | 84 | All SZGR 2.0 genes in this pathway |
| CHEBOTAEV GR TARGETS DN | 120 | 73 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN UP | 612 | 367 | All SZGR 2.0 genes in this pathway |
| PEREZ TP63 TARGETS | 355 | 243 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 AND TP63 TARGETS | 205 | 145 | All SZGR 2.0 genes in this pathway |
| LIANG HEMATOPOIESIS STEM CELL NUMBER LARGE VS TINY DN | 45 | 24 | All SZGR 2.0 genes in this pathway |
| LIANG HEMATOPOIESIS STEM CELL NUMBER SMALL VS HUGE DN | 33 | 22 | All SZGR 2.0 genes in this pathway |
| SCHLESINGER METHYLATED DE NOVO IN CANCER | 88 | 64 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA HIF1A DN | 110 | 78 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| JI METASTASIS REPRESSED BY STK11 | 27 | 17 | All SZGR 2.0 genes in this pathway |
| SWEET KRAS TARGETS UP | 84 | 51 | All SZGR 2.0 genes in this pathway |
| NADLER HYPERGLYCEMIA AT OBESITY | 58 | 35 | All SZGR 2.0 genes in this pathway |
| PETROVA ENDOTHELIUM LYMPHATIC VS BLOOD DN | 162 | 102 | All SZGR 2.0 genes in this pathway |
| MODY HIPPOCAMPUS POSTNATAL | 63 | 50 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS DN | 371 | 218 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR DN | 298 | 200 | All SZGR 2.0 genes in this pathway |
| MCCLUNG DELTA FOSB TARGETS 2WK | 48 | 36 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION DN | 287 | 208 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 1 | 419 | 273 | All SZGR 2.0 genes in this pathway |
| JIANG AGING CEREBRAL CORTEX DN | 54 | 43 | All SZGR 2.0 genes in this pathway |
| DURCHDEWALD SKIN CARCINOGENESIS DN | 264 | 168 | All SZGR 2.0 genes in this pathway |
| LIN NPAS4 TARGETS DN | 68 | 48 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB DN | 342 | 220 | All SZGR 2.0 genes in this pathway |
| LIANG HEMATOPOIESIS STEM CELL NUMBER QTL | 17 | 10 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 2 | 86 | 50 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 2HR DN | 49 | 33 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT | 567 | 365 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS UP | 279 | 155 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-1/206 | 3262 | 3268 | 1A | hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA |
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| miR-101 | 2958 | 2964 | 1A | hsa-miR-101 | UACAGUACUGUGAUAACUGAAG |
| miR-128 | 2624 | 2630 | m8 | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-130/301 | 2734 | 2740 | m8 | hsa-miR-130abrain | CAGUGCAAUGUUAAAAGGGCAU |
| hsa-miR-301 | CAGUGCAAUAGUAUUGUCAAAGC | ||||
| hsa-miR-130bbrain | CAGUGCAAUGAUGAAAGGGCAU | ||||
| hsa-miR-454-3p | UAGUGCAAUAUUGCUUAUAGGGUUU | ||||
| hsa-miR-130abrain | CAGUGCAAUGUUAAAAGGGCAU | ||||
| hsa-miR-301 | CAGUGCAAUAGUAUUGUCAAAGC | ||||
| hsa-miR-130bbrain | CAGUGCAAUGAUGAAAGGGCAU | ||||
| hsa-miR-454-3p | UAGUGCAAUAUUGCUUAUAGGGUUU | ||||
| miR-133 | 3311 | 3317 | m8 | hsa-miR-133a | UUGGUCCCCUUCAACCAGCUGU |
| hsa-miR-133b | UUGGUCCCCUUCAACCAGCUA | ||||
| miR-138 | 3183 | 3189 | m8 | hsa-miR-138brain | AGCUGGUGUUGUGAAUC |
| miR-139 | 2959 | 2966 | 1A,m8 | hsa-miR-139brain | UCUACAGUGCACGUGUCU |
| miR-144 | 2958 | 2964 | 1A | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| miR-148/152 | 2735 | 2742 | 1A,m8 | hsa-miR-148a | UCAGUGCACUACAGAACUUUGU |
| hsa-miR-152brain | UCAGUGCAUGACAGAACUUGGG | ||||
| hsa-miR-148b | UCAGUGCAUCACAGAACUUUGU | ||||
| miR-182 | 3661 | 3668 | 1A,m8 | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| miR-200bc/429 | 2943 | 2950 | 1A,m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-204/211 | 3597 | 3604 | 1A,m8 | hsa-miR-204brain | UUCCCUUUGUCAUCCUAUGCCU |
| hsa-miR-211 | UUCCCUUUGUCAUCCUUCGCCU | ||||
| miR-210 | 127 | 133 | m8 | hsa-miR-210 | CUGUGCGUGUGACAGCGGCUGA |
| miR-25/32/92/363/367 | 3584 | 3590 | m8 | hsa-miR-25brain | CAUUGCACUUGUCUCGGUCUGA |
| hsa-miR-32 | UAUUGCACAUUACUAAGUUGC | ||||
| hsa-miR-92 | UAUUGCACUUGUCCCGGCCUG | ||||
| hsa-miR-367 | AAUUGCACUUUAGCAAUGGUGA | ||||
| hsa-miR-92bSZ | UAUUGCACUCGUCCCGGCCUC | ||||
| miR-34/449 | 1600 | 1606 | m8 | hsa-miR-34abrain | UGGCAGUGUCUUAGCUGGUUGUU |
| hsa-miR-34c | AGGCAGUGUAGUUAGCUGAUUGC | ||||
| hsa-miR-449 | UGGCAGUGUAUUGUUAGCUGGU | ||||
| hsa-miR-449b | AGGCAGUGUAUUGUUAGCUGGC | ||||
| miR-346 | 1623 | 1629 | 1A | hsa-miR-346brain | UGUCUGCCCGCAUGCCUGCCUCU |
| miR-34b | 1601 | 1607 | m8 | hsa-miR-34b | UAGGCAGUGUCAUUAGCUGAUUG |
| miR-369-3p | 2945 | 2951 | m8 | hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU |
| miR-539 | 2950 | 2956 | m8 | hsa-miR-539 | GGAGAAAUUAUCCUUGGUGUGU |
| miR-9 | 3776 | 3782 | m8 | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
| miR-96 | 3662 | 3668 | 1A | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.