Gene Page: NUP98
Summary ?
| GeneID | 4928 |
| Symbol | NUP98 |
| Synonyms | ADIR2|NUP196|NUP96 |
| Description | nucleoporin 98kDa |
| Reference | MIM:601021|HGNC:HGNC:8068|Ensembl:ENSG00000110713|HPRD:03012|Vega:OTTHUMG00000011846 |
| Gene type | protein-coding |
| Map location | 11p15.5 |
| Pascal p-value | 0.152 |
| Fetal beta | 0.39 |
| eGene | Caudate basal ganglia |
| Support | CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:McCarthy_2014 | Whole Exome Sequencing analysis | Whole exome sequencing of 57 trios with sporadic or familial schizophrenia. |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NUP98 | chr11 | 3697753 | G | C | NM_139132 | p.R1650P | missense SNV | C | P | Schizophrenia | DNM:McCarthy_2014 |
Section II. Transcriptome annotation
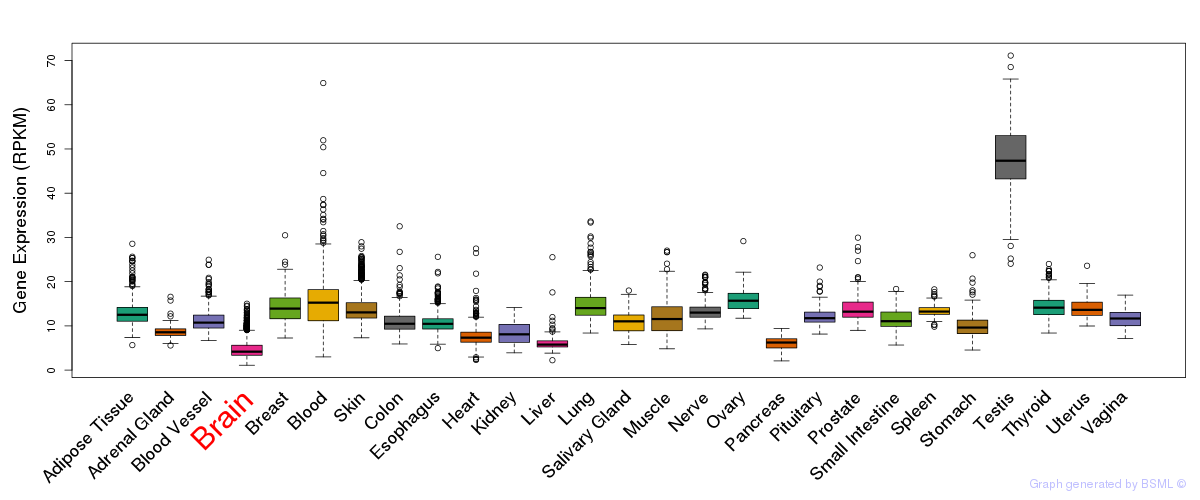
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | - | HPRD,BioGRID | 9858599 |
| EP300 | KAT3B | p300 | E1A binding protein p300 | Affinity Capture-Western | BioGRID | 9858599 |
| IPO5 | DKFZp686O1576 | FLJ43041 | IMB3 | KPNB3 | MGC2068 | RANBP5 | importin 5 | - | HPRD | 9114010 |
| KPNB1 | IMB1 | IPOB | Impnb | MGC2155 | MGC2156 | MGC2157 | NTF97 | karyopherin (importin) beta 1 | - | HPRD,BioGRID | 9144189 |
| MOBKL1B | C2orf6 | FLJ10788 | FLJ11595 | MATS1 | MOB1 | MOBK1B | Mob4B | MOB1, Mps One Binder kinase activator-like 1B (yeast) | Affinity Capture-MS | BioGRID | 17353931 |
| NFX1 | DKFZp779G2416 | MGC20369 | NFX2 | nuclear transcription factor, X-box binding 1 | Reconstituted Complex | BioGRID | 11073998 |
| NUP133 | FLJ10814 | MGC21133 | hNUP133 | nucleoporin 133kDa | - | HPRD,BioGRID | 11564755 |
| NUP214 | CAIN | CAN | D9S46E | MGC104525 | N214 | nucleoporin 214kDa | - | HPRD | 12589057 |
| NUP62 | DKFZp547L134 | FLJ20822 | FLJ43869 | IBSN | MGC841 | SNDI | p62 | nucleoporin 62kDa | - | HPRD | 8707840 |
| NUP88 | MGC8530 | nucleoporin 88kDa | - | HPRD,BioGRID | 12589057 |
| NUP98 | ADIR2 | NUP196 | NUP96 | nucleoporin 98kDa | - | HPRD | 12589057 |
| NXF1 | DKFZp667O0311 | MEX67 | TAP | nuclear RNA export factor 1 | - | HPRD | 10668806 |
| NXF1 | DKFZp667O0311 | MEX67 | TAP | nuclear RNA export factor 1 | Reconstituted Complex | BioGRID | 11551912 |
| PARP11 | C12orf6 | DKFZp779H0122 | poly (ADP-ribose) polymerase family, member 11 | Affinity Capture-MS | BioGRID | 17353931 |
| QRICH2 | DKFZp434P0316 | glutamine rich 2 | Affinity Capture-MS | BioGRID | 17353931 |
| RAE1 | FLJ30608 | MGC117333 | MGC126076 | MGC126077 | MIG14 | MRNP41 | Mnrp41 | dJ481F12.3 | dJ800J21.1 | RAE1 RNA export 1 homolog (S. pombe) | - | HPRD,BioGRID | 10209021 |
| TNPO1 | IPO2 | KPNB2 | MIP | MIP1 | TRN | transportin 1 | - | HPRD,BioGRID | 9144189|10875935 |
| TNPO2 | FLJ12155 | IPO3 | KPNB2B | TRN2 | transportin 2 (importin 3, karyopherin beta 2b) | Reconstituted Complex | BioGRID | 10875935 |
| TPR | - | translocated promoter region (to activated MET oncogene) | - | HPRD,BioGRID | 11248057 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| HORIUCHI WTAP TARGETS DN | 310 | 188 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA DN | 146 | 94 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA BY DMOG DN | 59 | 40 | All SZGR 2.0 genes in this pathway |
| KLEIN TARGETS OF BCR ABL1 FUSION | 45 | 34 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER MYC TARGETS REPRESSED BY SERUM | 159 | 93 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 1 DN | 169 | 102 | All SZGR 2.0 genes in this pathway |
| PENG LEUCINE DEPRIVATION DN | 187 | 122 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA DN | 289 | 166 | All SZGR 2.0 genes in this pathway |
| CHEOK RESPONSE TO HD MTX DN | 24 | 18 | All SZGR 2.0 genes in this pathway |
| PENG GLUCOSE DEPRIVATION DN | 169 | 112 | All SZGR 2.0 genes in this pathway |
| PENG RAPAMYCIN RESPONSE DN | 245 | 154 | All SZGR 2.0 genes in this pathway |
| PENG GLUTAMINE DEPRIVATION DN | 337 | 230 | All SZGR 2.0 genes in this pathway |
| HASLINGER B CLL WITH 6Q21 DELETION | 20 | 15 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT UP | 390 | 242 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION UP | 314 | 201 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR UP | 178 | 111 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION DN | 287 | 208 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 24HR UP | 148 | 96 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE DN | 373 | 196 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| TOOKER GEMCITABINE RESISTANCE UP | 79 | 40 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS DN | 366 | 257 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G6 DN | 19 | 13 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE G2 M | 216 | 124 | All SZGR 2.0 genes in this pathway |
| RATTENBACHER BOUND BY CELF1 | 467 | 251 | All SZGR 2.0 genes in this pathway |