Gene Page: PCSK1
Summary ?
| GeneID | 5122 |
| Symbol | PCSK1 |
| Synonyms | BMIQ12|NEC1|PC1|PC3|SPC3 |
| Description | proprotein convertase subtilisin/kexin type 1 |
| Reference | MIM:162150|HGNC:HGNC:8743|Ensembl:ENSG00000175426|HPRD:01201|Vega:OTTHUMG00000122089 |
| Gene type | protein-coding |
| Map location | 5q15-q21 |
| Pascal p-value | 0.01 |
| Sherlock p-value | 0.223 |
| Fetal beta | -4.828 |
| eGene | Myers' cis & trans |
| Support | NEUROTROPHIN SIGNALING |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenics,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1467642 | chr10 | 21058685 | PCSK1 | 5122 | 0.17 | trans | ||
| rs8036059 | chr15 | 28112722 | PCSK1 | 5122 | 0.19 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
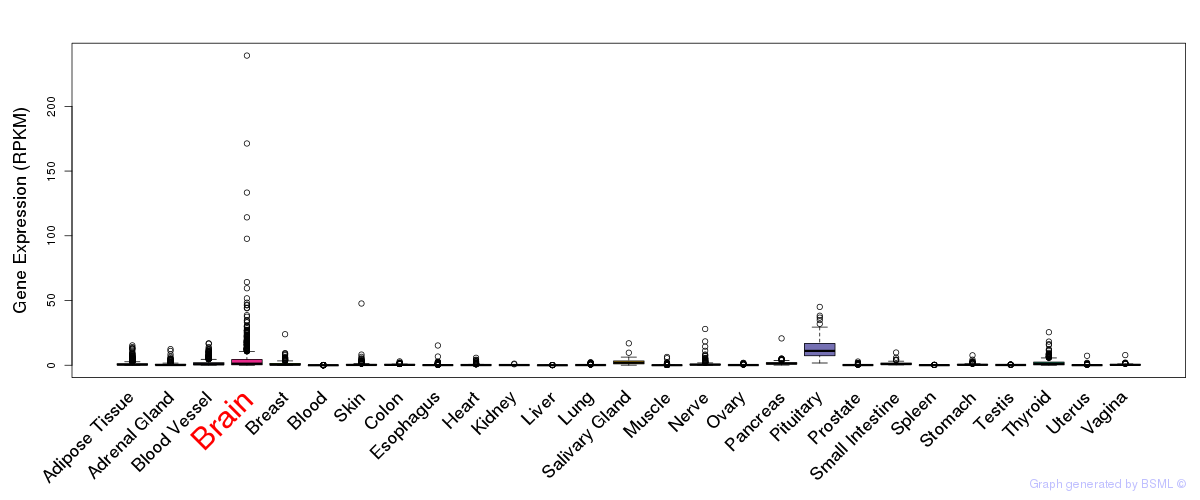
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| BIN1 | 0.81 | 0.82 |
| PIGZ | 0.77 | 0.81 |
| KIFC2 | 0.76 | 0.82 |
| SYNGR3 | 0.76 | 0.79 |
| FBXO27 | 0.75 | 0.76 |
| P2RX5 | 0.75 | 0.77 |
| RHOF | 0.73 | 0.74 |
| C19orf28 | 0.73 | 0.74 |
| KCNQ4 | 0.73 | 0.78 |
| AMDHD2 | 0.73 | 0.73 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.41 | -0.19 |
| RPS20 | -0.41 | -0.46 |
| SNRPG | -0.38 | -0.41 |
| TPT1 | -0.38 | -0.35 |
| CLIC1 | -0.36 | -0.45 |
| RAB13 | -0.36 | -0.37 |
| FAM60A | -0.36 | -0.17 |
| RPL24 | -0.36 | -0.35 |
| NCAPG | -0.36 | -0.15 |
| RPL27 | -0.35 | -0.35 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004252 | serine-type endopeptidase activity | IEA | glutamate (GO term level: 7) | - |
| GO:0005509 | calcium ion binding | IEA | - | |
| GO:0008233 | peptidase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006508 | proteolysis | TAS | 9207799 | |
| GO:0007267 | cell-cell signaling | TAS | 9207799 | |
| GO:0008152 | metabolic process | TAS | 9207799 | |
| GO:0043043 | peptide biosynthetic process | ISS | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005615 | extracellular space | IDA | 8397508 | |
| GO:0031410 | cytoplasmic vesicle | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | 200 | 136 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRATION OF ENERGY METABOLISM | 120 | 84 | All SZGR 2.0 genes in this pathway |
| REACTOME DIABETES PATHWAYS | 133 | 91 | All SZGR 2.0 genes in this pathway |
| REACTOME PEPTIDE HORMONE BIOSYNTHESIS | 14 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME INSULIN SYNTHESIS AND PROCESSING | 21 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF INSULIN SECRETION | 93 | 65 | All SZGR 2.0 genes in this pathway |
| REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | 16 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | 14 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME INCRETIN SYNTHESIS SECRETION AND INACTIVATION | 22 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | 19 | 12 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| SABATES COLORECTAL ADENOMA UP | 141 | 75 | All SZGR 2.0 genes in this pathway |
| CONCANNON APOPTOSIS BY EPOXOMICIN UP | 239 | 157 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF ICP WITH H3K4ME3 AND H3K27ME3 | 38 | 34 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 | 718 | 401 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| SERVITJA ISLET HNF1A TARGETS DN | 109 | 71 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS UP | 279 | 155 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-186 | 2293 | 2299 | 1A | hsa-miR-186 | CAAAGAAUUCUCCUUUUGGGCUU |
| miR-203.1 | 309 | 315 | 1A | hsa-miR-203 | UGAAAUGUUUAGGACCACUAG |
| miR-320 | 2129 | 2135 | 1A | hsa-miR-320 | AAAAGCUGGGUUGAGAGGGCGAA |
| miR-410 | 2257 | 2263 | 1A | hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
| miR-494 | 2159 | 2165 | m8 | hsa-miR-494brain | UGAAACAUACACGGGAAACCUCUU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.