Gene Page: PFN1
Summary ?
| GeneID | 5216 |
| Symbol | PFN1 |
| Synonyms | ALS18 |
| Description | profilin 1 |
| Reference | MIM:176610|HGNC:HGNC:8881|Ensembl:ENSG00000108518|HPRD:01452|Vega:OTTHUMG00000099396 |
| Gene type | protein-coding |
| Map location | 17p13.3 |
| Pascal p-value | 0.75 |
| Sherlock p-value | 0.516 |
| Fetal beta | 0.535 |
| DMG | 1 (# studies) |
| Support | STRUCTURAL PLASTICITY G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 1 |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.2331 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg20693880 | 17 | 4699478 | PFN1 | 0.002 | 2.552 | DMG:vanEijk_2014 |
Section II. Transcriptome annotation
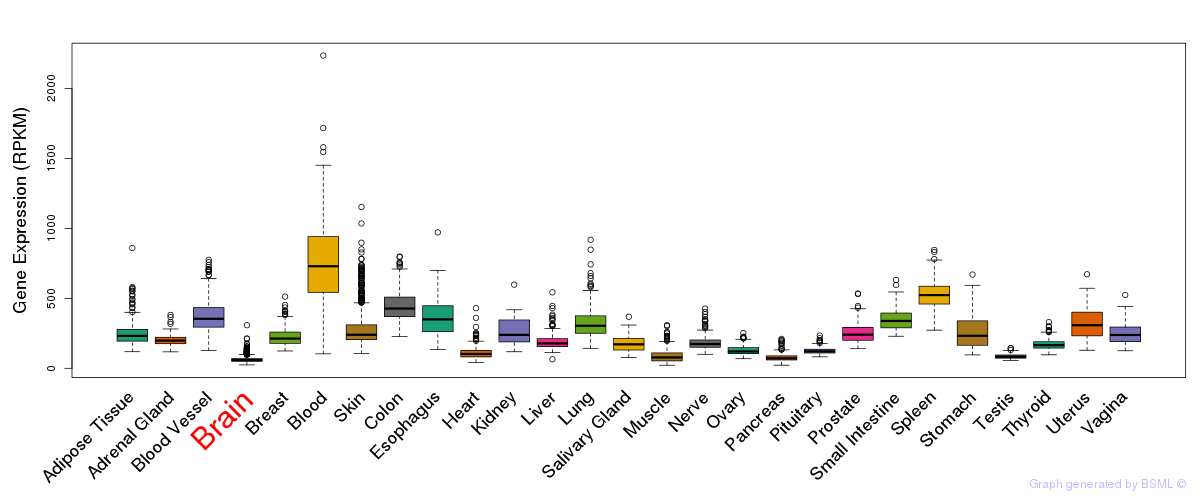
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003779 | actin binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007010 | cytoskeleton organization | IEA | - | |
| GO:0030036 | actin cytoskeleton organization | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0015629 | actin cytoskeleton | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTA1 | ACTA | ASMA | CFTD | CFTD1 | CFTDM | MPFD | NEM1 | NEM2 | NEM3 | actin, alpha 1, skeletal muscle | - | HPRD | 8413665 |
| ACTB | PS1TP5BP1 | actin, beta | - | HPRD,BioGRID | 10411937 |
| APBB1IP | INAG1 | PREL1 | RARP1 | RIAM | amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein | RIAM interacts with Profilin. | BIND | 15469846 |
| APBB1IP | INAG1 | PREL1 | RARP1 | RIAM | amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein | - | HPRD | 15469846 |
| C14orf1 | ERG28 | NET51 | chromosome 14 open reading frame 1 | Two-hybrid | BioGRID | 16169070 |
| C1orf103 | FLJ11269 | RIF1 | RP11-96K19.1 | chromosome 1 open reading frame 103 | Two-hybrid | BioGRID | 16169070 |
| CRMP1 | DPYSL1 | DRP-1 | DRP1 | collapsin response mediator protein 1 | Two-hybrid | BioGRID | 16169070 |
| DLG5 | KIAA0583 | LP-DLG | P-DLG5 | PDLG | discs, large homolog 5 (Drosophila) | Two-hybrid | BioGRID | 16169070 |
| DNM2 | CMTDI1 | CMTDIB | DI-CMTB | DYN2 | DYNII | dynamin 2 | - | HPRD | 12419186 |
| ENAH | ENA | MENA | NDPP1 | enabled homolog (Drosophila) | - | HPRD,BioGRID | 9473484 |
| FMNL1 | C17orf1 | C17orf1B | FHOD4 | FMNL | KW-13 | MGC133052 | MGC1894 | MGC21878 | formin-like 1 | - | HPRD,BioGRID | 10958683 |
| GPHN | GEPH | GPH | GPHRYN | KIAA1385 | gephyrin | - | HPRD,BioGRID | 9473484 |
| LOC339344 | - | hypothetical protein LOC339344 | - | HPRD | 15615774 |
| MLLT4 | AF-6 | AF6 | AFADIN | FLJ34371 | RP3-431P23.3 | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 4 | - | HPRD,BioGRID | 10922060 |
| NCK1 | MGC12668 | NCK | NCKalpha | NCK adaptor protein 1 | Reconstituted Complex | BioGRID | 9694849 |
| RHOQ | ARHQ | RASL7A | TC10 | TC10A | ras homolog gene family, member Q | - | HPRD,BioGRID | 10445846 |
| TLE1 | ESG | ESG1 | GRG1 | transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila) | Two-hybrid | BioGRID | 16169070 |
| UNC119 | HRG4 | unc-119 homolog (C. elegans) | Two-hybrid | BioGRID | 16169070 |
| VASP | - | vasodilator-stimulated phosphoprotein | - | HPRD,BioGRID | 10882740 |
| VIPR1 | FLJ41949 | HVR1 | II | PACAP-R-2 | RDC1 | VAPC1 | VIPR | VIRG | VPAC1 | VPCAP1R | vasoactive intestinal peptide receptor 1 | Reconstituted Complex | BioGRID | 10867004 |
| WASF1 | FLJ31482 | KIAA0269 | SCAR1 | WAVE | WAVE1 | WAS protein family, member 1 | - | HPRD,BioGRID | 9843499 |
| WASL | DKFZp779G0847 | MGC48327 | N-WASP | NWASP | Wiskott-Aldrich syndrome-like | - | HPRD,BioGRID | 9822597 |
| WIPF2 | WICH | WIRE | WAS/WASL interacting protein family, member 2 | - | HPRD | 12213210 |
| XPO6 | EXP6 | FLJ22519 | KIAA0370 | RANBP20 | exportin 6 | - | HPRD,BioGRID | 14592989 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG REGULATION OF ACTIN CYTOSKELETON | 216 | 144 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ECM PATHWAY | 24 | 17 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RHO PATHWAY | 32 | 23 | All SZGR 2.0 genes in this pathway |
| SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | 35 | 25 | All SZGR 2.0 genes in this pathway |
| ST FAS SIGNALING PATHWAY | 65 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | 89 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME AXON GUIDANCE | 251 | 188 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ROBO RECEPTOR | 30 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | 208 | 138 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES SKIN UP | 177 | 113 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| WANG BARRETTS ESOPHAGUS UP | 51 | 27 | All SZGR 2.0 genes in this pathway |
| MATTIOLI MGUS VS PCL | 116 | 62 | All SZGR 2.0 genes in this pathway |
| DAIRKEE TERT TARGETS UP | 380 | 213 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| GOTZMANN EPITHELIAL TO MESENCHYMAL TRANSITION DN | 206 | 136 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| SAKAI TUMOR INFILTRATING MONOCYTES UP | 27 | 19 | All SZGR 2.0 genes in this pathway |
| ZUCCHI METASTASIS DN | 44 | 29 | All SZGR 2.0 genes in this pathway |
| LIN APC TARGETS | 77 | 55 | All SZGR 2.0 genes in this pathway |
| PENG GLUTAMINE DEPRIVATION DN | 337 | 230 | All SZGR 2.0 genes in this pathway |
| NADLER OBESITY UP | 61 | 34 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN MGMT 24HR DN | 33 | 21 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK UP | 244 | 151 | All SZGR 2.0 genes in this pathway |
| DAZARD UV RESPONSE CLUSTER G4 | 21 | 15 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY UP | 487 | 303 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN WITHOUT MGMT 48HR DN | 32 | 25 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN MGMT 48HR DN | 161 | 105 | All SZGR 2.0 genes in this pathway |
| JIANG AGING CEREBRAL CORTEX DN | 54 | 43 | All SZGR 2.0 genes in this pathway |
| DURCHDEWALD SKIN CARCINOGENESIS DN | 264 | 168 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY GAMMA RADIATION | 81 | 59 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE UP | 226 | 164 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| QI PLASMACYTOMA UP | 259 | 185 | All SZGR 2.0 genes in this pathway |
| ALONSO METASTASIS EMT UP | 36 | 24 | All SZGR 2.0 genes in this pathway |
| ALONSO METASTASIS UP | 198 | 128 | All SZGR 2.0 genes in this pathway |
| MUELLER PLURINET | 299 | 189 | All SZGR 2.0 genes in this pathway |
| CHANG CORE SERUM RESPONSE UP | 212 | 128 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| FOURNIER ACINAR DEVELOPMENT LATE 2 | 277 | 172 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S1 | 237 | 159 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| KYNG RESPONSE TO H2O2 VIA ERCC6 DN | 46 | 31 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 UP | 344 | 215 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 SIGNALING VIA NFIC 1HR DN | 106 | 77 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-140 | 117 | 124 | 1A,m8 | hsa-miR-140brain | AGUGGUUUUACCCUAUGGUAG |
| miR-182 | 112 | 118 | 1A | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| miR-19 | 38 | 44 | m8 | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-96 | 112 | 118 | 1A | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.