Gene Page: CLIC5
Summary ?
| GeneID | 53405 |
| Symbol | CLIC5 |
| Synonyms | DFNB102|DFNB103|MST130|MSTP130 |
| Description | chloride intracellular channel 5 |
| Reference | MIM:607293|HGNC:HGNC:13517|Ensembl:ENSG00000112782|HPRD:06292|Vega:OTTHUMG00000014775 |
| Gene type | protein-coding |
| Map location | 6p12.3 |
| Pascal p-value | 0.129 |
| Fetal beta | 0.302 |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Xu_2012 | Whole Exome Sequencing analysis | De novo mutations of 4 genes were identified by exome sequencing of 795 samples in this study | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.04433 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CLIC5 | chr6 | 46047670 | C | A | NM_001114086 | p.104G>C | missense | Schizophrenia | DNM:Xu_2012 |
Section II. Transcriptome annotation
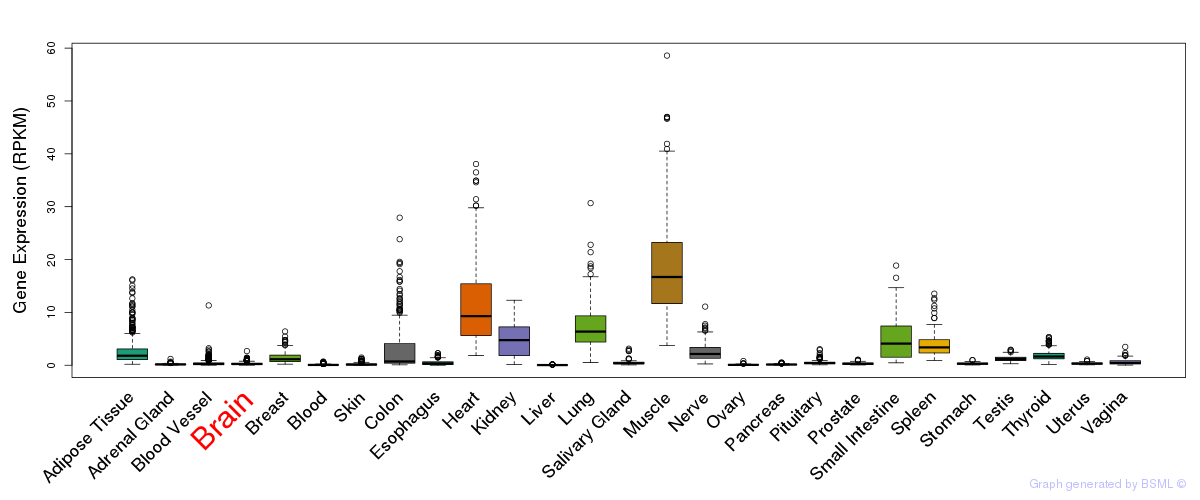
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 10793131 | |
| GO:0005244 | voltage-gated ion channel activity | IEA | - | |
| GO:0005247 | voltage-gated chloride channel activity | IEA | - | |
| GO:0031404 | chloride ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008104 | protein localization | IEA | - | |
| GO:0007605 | sensory perception of sound | IEA | - | |
| GO:0007565 | female pregnancy | TAS | 10793131 | |
| GO:0006821 | chloride transport | IDA | 10793131 | |
| GO:0006811 | ion transport | IEA | - | |
| GO:0050885 | neuromuscular process controlling balance | IEA | - | |
| GO:0060088 | auditory receptor cell stereocilium organization | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005794 | Golgi apparatus | IEA | - | |
| GO:0005813 | centrosome | IEA | - | |
| GO:0005626 | insoluble fraction | IDA | 10793131 | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0016020 | membrane | IEA | - | |
| GO:0015629 | actin cytoskeleton | IDA | 10793131 | |
| GO:0032420 | stereocilium | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| THUM SYSTOLIC HEART FAILURE UP | 423 | 283 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER GRADES 1 2 DN | 67 | 43 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS DN | 31 | 23 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL DN | 216 | 143 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS HCP WITH H3 UNMETHYLATED | 80 | 50 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| SERVITJA ISLET HNF1A TARGETS DN | 109 | 71 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL DN | 308 | 187 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-122 | 4419 | 4426 | 1A,m8 | hsa-miR-122a | UGGAGUGUGACAAUGGUGUUUGU |
| miR-182 | 761 | 767 | 1A | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| miR-26 | 1247 | 1253 | 1A | hsa-miR-26abrain | UUCAAGUAAUCCAGGAUAGGC |
| hsa-miR-26bSZ | UUCAAGUAAUUCAGGAUAGGUU | ||||
| miR-96 | 761 | 767 | 1A | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.