Gene Page: POU2F1
Summary ?
| GeneID | 5451 |
| Symbol | POU2F1 |
| Synonyms | OCT1|OTF1|oct-1B |
| Description | POU class 2 homeobox 1 |
| Reference | MIM:164175|HGNC:HGNC:9212|Ensembl:ENSG00000143190|HPRD:01250|Vega:OTTHUMG00000034436 |
| Gene type | protein-coding |
| Map location | 1q24.2 |
| Sherlock p-value | 0.906 |
| Fetal beta | 1.433 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg23087020 | 1 | 167190262 | POU2F1 | 2.57E-5 | 0.406 | 0.017 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17392743 | chr1 | 54764240 | POU2F1 | 5451 | 0.16 | trans | ||
| rs823371 | chr7 | 29648352 | POU2F1 | 5451 | 0.06 | trans | ||
| rs16955618 | chr15 | 29937543 | POU2F1 | 5451 | 0.07 | trans |
Section II. Transcriptome annotation
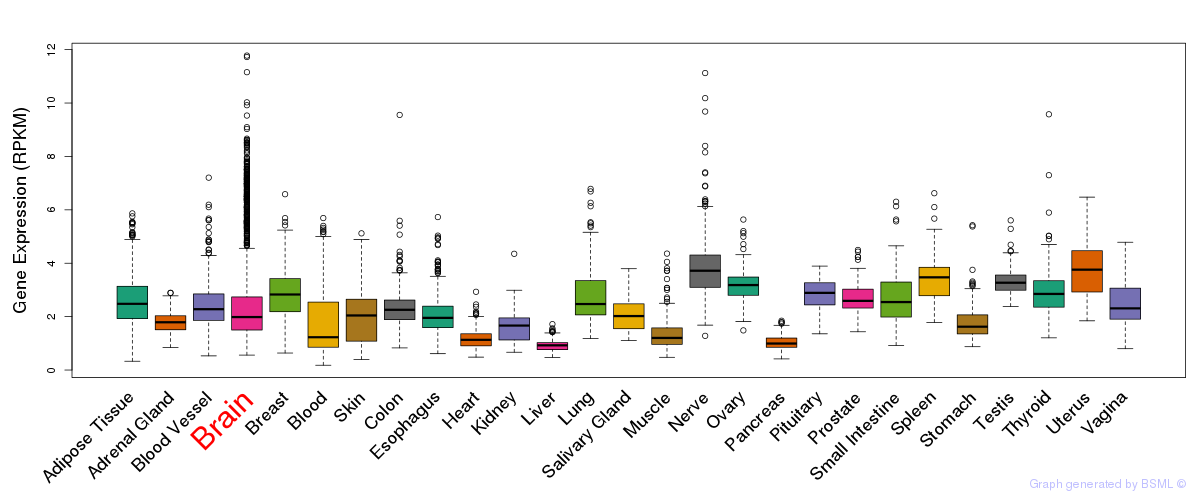
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | - | HPRD,BioGRID | 10480874 |
| BARD1 | - | BRCA1 associated RING domain 1 | - | HPRD | 11777930 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | Affinity Capture-Western | BioGRID | 11777930 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | BRCA1 interacts with Oct-1. | BIND | 11777930 |
| EPRS | DKFZp313B047 | EARS | GLUPRORS | PARS | PIG32 | QARS | QPRS | glutamyl-prolyl-tRNA synthetase | Far Western Reconstituted Complex Two-hybrid | BioGRID | 9537509 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | Reconstituted Complex | BioGRID | 10480874 |
| FDXR | ADXR | ferredoxin reductase | - | HPRD | 11096094 |
| GAPDH | G3PD | GAPD | MGC88685 | glyceraldehyde-3-phosphate dehydrogenase | Affinity Capture-Western Reconstituted Complex | BioGRID | 12887926 |
| HCFC1 | CFF | HCF-1 | HCF1 | HFC1 | MGC70925 | VCAF | host cell factor C1 (VP16-accessory protein) | - | HPRD,BioGRID | 8454622 |
| HMGB2 | HMG2 | high-mobility group box 2 | - | HPRD | 7720710 |
| HOXC4 | HOX3 | HOX3E | cp19 | homeobox C4 | - | HPRD | 12672812 |
| LIFR | CD118 | FLJ98106 | FLJ99923 | LIF-R | SJS2 | STWS | SWS | leukemia inhibitory factor receptor alpha | - | HPRD | 10383413 |
| MNAT1 | MAT1 | RNF66 | TFB3 | menage a trois homolog 1, cyclin H assembly factor (Xenopus laevis) | - | HPRD,BioGRID | 9368058 |
| NCOR2 | CTG26 | SMRT | SMRTE | SMRTE-tau | TNRC14 | TRAC-1 | TRAC1 | nuclear receptor co-repressor 2 | - | HPRD,BioGRID | 11134019 |
| NFYA | CBF-A | CBF-B | FLJ11236 | HAP2 | NF-YA | nuclear transcription factor Y, alpha | - | HPRD | 11777930 |
| NFYA | CBF-A | CBF-B | FLJ11236 | HAP2 | NF-YA | nuclear transcription factor Y, alpha | Oct-1 interacts with NF-YA | BIND | 11777930 |
| NPAT | E14 | nuclear protein, ataxia-telangiectasia locus | - | HPRD,BioGRID | 12887926 |
| NR3C1 | GCCR | GCR | GR | GRL | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) | - | HPRD,BioGRID | 9584182 |10480874 |10490647 |14522952 |
| PARP1 | ADPRT | ADPRT1 | PARP | PARP-1 | PPOL | pADPRT-1 | poly (ADP-ribose) polymerase 1 | - | HPRD | 9537509 |
| PGR | NR3C3 | PR | progesterone receptor | Reconstituted Complex | BioGRID | 10480874 |
| POU2AF1 | BOB1 | OBF-1 | OBF1 | OCAB | POU class 2 associating factor 1 | - | HPRD | 11380252 |12727885 |
| POU2AF1 | BOB1 | OBF-1 | OBF1 | OCAB | POU class 2 associating factor 1 | Co-crystal Structure Reconstituted Complex Two-hybrid | BioGRID | 9418884 |10541551 |11380252 |
| POU2F1 | OCT1 | OTF1 | POU class 2 homeobox 1 | Co-crystal Structure | BioGRID | 11583619 |
| RARA | NR1B1 | RAR | retinoic acid receptor, alpha | Reconstituted Complex | BioGRID | 10480874 |
| RELA | MGC131774 | NFKB3 | p65 | v-rel reticuloendotheliosis viral oncogene homolog A (avian) | - | HPRD,BioGRID | 12019209 |
| RXRA | FLJ00280 | FLJ00318 | FLJ16020 | FLJ16733 | MGC102720 | NR2B1 | retinoid X receptor, alpha | - | HPRD,BioGRID | 10383413 |
| SNAPC4 | FLJ13451 | PTFalpha | SNAP190 | small nuclear RNA activating complex, polypeptide 4, 190kDa | - | HPRD,BioGRID | 9418884 |11856838 |
| SOX2 | ANOP3 | MCOPS3 | MGC2413 | SRY (sex determining region Y)-box 2 | - | HPRD | 12923055 |
| SP1 | - | Sp1 transcription factor | - | HPRD,BioGRID | 8668525 |
| STIP1 | HOP | IEF-SSP-3521 | P60 | STI1 | STI1L | stress-induced-phosphoprotein 1 | Affinity Capture-Western | BioGRID | 12887926 |
| TBP | GTF2D | GTF2D1 | MGC117320 | MGC126054 | MGC126055 | SCA17 | TFIID | TATA box binding protein | - | HPRD,BioGRID | 8202368 |
| THRA | AR7 | EAR7 | ERB-T-1 | ERBA | ERBA1 | MGC000261 | MGC43240 | NR1A1 | THRA1 | THRA2 | c-ERBA-1 | thyroid hormone receptor, alpha (erythroblastic leukemia viral (v-erb-a) oncogene homolog, avian) | Reconstituted Complex | BioGRID | 10480874 |
| TNF | DIF | TNF-alpha | TNFA | TNFSF2 | tumor necrosis factor (TNF superfamily, member 2) | Reconstituted Complex | BioGRID | 12019209 |
| TXNRD1 | GRIM-12 | MGC9145 | TR | TR1 | TRXR1 | TXNR | thioredoxin reductase 1 | Reconstituted Complex | BioGRID | 10383413 |
| VDR | NR1I1 | vitamin D (1,25- dihydroxyvitamin D3) receptor | Reconstituted Complex | BioGRID | 10383413 |
| XRCC5 | FLJ39089 | KARP-1 | KARP1 | KU80 | KUB2 | Ku86 | NFIV | X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining) | Affinity Capture-Western | BioGRID | 11279128 |12393188 |
| XRCC6 | CTC75 | CTCBF | G22P1 | KU70 | ML8 | TLAA | X-ray repair complementing defective repair in Chinese hamster cells 6 | Affinity Capture-Western | BioGRID | 11279128 |
| XRCC6 | CTC75 | CTCBF | G22P1 | KU70 | ML8 | TLAA | X-ray repair complementing defective repair in Chinese hamster cells 6 | - | HPRD | 12672812 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID NFAT TFPATHWAY | 47 | 39 | All SZGR 2.0 genes in this pathway |
| PID REG GR PATHWAY | 82 | 60 | All SZGR 2.0 genes in this pathway |
| PID AR TF PATHWAY | 53 | 38 | All SZGR 2.0 genes in this pathway |
| PID ATF2 PATHWAY | 59 | 43 | All SZGR 2.0 genes in this pathway |
| PID TCR CALCIUM PATHWAY | 29 | 23 | All SZGR 2.0 genes in this pathway |
| PID HNF3A PATHWAY | 44 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL III TRANSCRIPTION | 33 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTION | 210 | 127 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | 122 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | 26 | 13 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL DN | 275 | 168 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION | 77 | 51 | All SZGR 2.0 genes in this pathway |
| DAIRKEE TERT TARGETS DN | 124 | 79 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| COLLIS PRKDC SUBSTRATES | 20 | 15 | All SZGR 2.0 genes in this pathway |
| CHESLER BRAIN QTL CIS | 75 | 51 | All SZGR 2.0 genes in this pathway |
| GUO HEX TARGETS UP | 81 | 54 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 16HR UP | 225 | 139 | All SZGR 2.0 genes in this pathway |
| MCCLUNG COCAINE REWARD 5D | 79 | 62 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 10HR UP | 101 | 69 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 24HR UP | 148 | 96 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY OVERCONNECTED IN BREAST CANCER | 22 | 19 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 36HR DN | 185 | 116 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS UP | 217 | 131 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS GROUP1 | 136 | 76 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| FOSTER KDM1A TARGETS DN | 211 | 119 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM A | 182 | 108 | All SZGR 2.0 genes in this pathway |