Gene Page: EXOSC4
Summary ?
| GeneID | 54512 |
| Symbol | EXOSC4 |
| Synonyms | RRP41|RRP41A|Rrp41p|SKI6|Ski6p|hRrp41p|p12A |
| Description | exosome component 4 |
| Reference | MIM:606491|HGNC:HGNC:18189|Ensembl:ENSG00000178896|HPRD:16221|Vega:OTTHUMG00000165437 |
| Gene type | protein-coding |
| Map location | 8q24.3 |
| Pascal p-value | 0.824 |
| Fetal beta | -0.251 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| Expression | Meta-analysis of gene expression | P value: 1.385 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg24028381 | 8 | 145133668 | EXOSC4 | 4.54E-5 | -0.301 | 0.021 | DMG:Wockner_2014 |
| cg25788012 | 8 | 145133899 | EXOSC4 | 1.398E-4 | -0.248 | 0.031 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
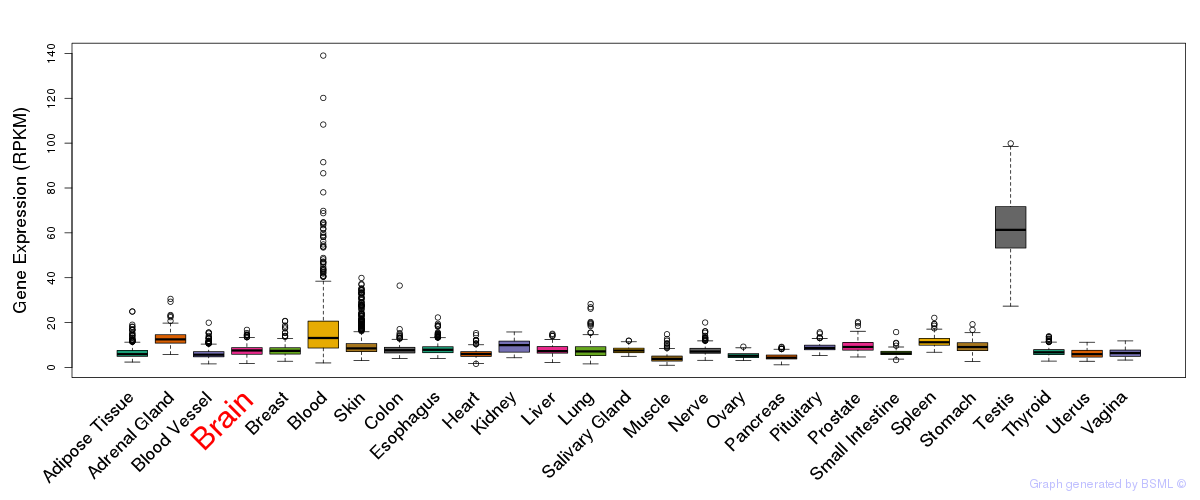
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000175 | 3'-5'-exoribonuclease activity | NAS | 11110791 | |
| GO:0003723 | RNA binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 15231747 | |
| GO:0004527 | exonuclease activity | IEA | - | |
| GO:0016787 | hydrolase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006396 | RNA processing | IEA | - | |
| GO:0006364 | rRNA processing | NAS | 11110791 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000178 | exosome (RNase complex) | IEA | - | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005730 | nucleolus | NAS | 11110791 | |
| GO:0005737 | cytoplasm | NAS | 11110791 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AKR1A1 | ALDR1 | ALR | ARM | DD3 | MGC12529 | MGC1380 | aldo-keto reductase family 1, member A1 (aldehyde reductase) | - | HPRD,BioGRID | 15231747 |
| DOM3Z | DOM3L | NG6 | dom-3 homolog Z (C. elegans) | - | HPRD | 15231747 |
| EEF1A1 | CCS-3 | CCS3 | EEF-1 | EEF1A | EF-Tu | EF1A | FLJ25721 | GRAF-1EF | HNGC:16303 | LENG7 | MGC102687 | MGC131894 | MGC16224 | PTI1 | eEF1A-1 | eukaryotic translation elongation factor 1 alpha 1 | - | HPRD,BioGRID | 15231747 |
| EXOSC1 | CGI-108 | CSL4 | Csl4p | SKI4 | Ski4p | hCsl4p | p13 | exosome component 1 | - | HPRD | 11110791 |12419256 |
| EXOSC1 | CGI-108 | CSL4 | Csl4p | SKI4 | Ski4p | hCsl4p | p13 | exosome component 1 | hCsl4p interacts with hRrp41p. | BIND | 12419256 |
| EXOSC10 | PM-Scl | PM/Scl-100 | PMSCL | PMSCL2 | RRP6 | Rrp6p | p2 | p3 | p4 | exosome component 10 | - | HPRD | 15231747 |
| EXOSC2 | RRP4 | Rrp4p | hRrp4p | p7 | exosome component 2 | - | HPRD | 12419256 |
| EXOSC2 | RRP4 | Rrp4p | hRrp4p | p7 | exosome component 2 | - | HPRD | 11719186 |
| EXOSC2 | RRP4 | Rrp4p | hRrp4p | p7 | exosome component 2 | hRrp4p interacts with hRrp41p. | BIND | 12419256 |
| EXOSC3 | CGI-102 | MGC15120 | MGC723 | RP11-3J10.8 | RRP40 | Rrp40p | bA3J10.7 | hRrp40p | p10 | exosome component 3 | - | HPRD | 11110791 |12419256 |
| EXOSC4 | FLJ20591 | RRP41 | RRP41A | Rrp41p | SKI6 | Ski6p | hRrp41p | p12A | exosome component 4 | hRrp41p interacts with another hRrp41p. | BIND | 12419256 |
| EXOSC4 | FLJ20591 | RRP41 | RRP41A | Rrp41p | SKI6 | Ski6p | hRrp41p | p12A | exosome component 4 | - | HPRD | 12419256 |
| EXOSC7 | EAP1 | FLJ26543 | KIAA0116 | RRP42 | Rrp42p | hRrp42p | p8 | exosome component 7 | hRrp42p interacts with hRrp41p. | BIND | 12419256 |
| EXOSC9 | PM/Scl-75 | PMSCL1 | RRP45 | Rrp45p | p5 | p6 | exosome component 9 | PM/Scl-75c-alpha interacts with hRrp41p. | BIND | 12788944 |
| GADD45GIP1 | CKBBP2 | CRIF1 | MGC4667 | MGC4758 | PLINP-1 | PRG6 | Plinp1 | growth arrest and DNA-damage-inducible, gamma interacting protein 1 | - | HPRD | 15231747 |
| GTF2IRD1 | BEN | CREAM1 | GTF3 | MUSTRD1 | RBAP2 | WBS | WBSCR11 | WBSCR12 | hMusTRD1alpha1 | GTF2I repeat domain containing 1 | - | HPRD,BioGRID | 15231747 |
| LRRC8D | FLJ10470 | FLJ20403 | LRRC5 | leucine rich repeat containing 8 family, member D | - | HPRD | 15231747 |
| MPZL1 | FLJ21047 | PZR | PZR1b | PZRa | PZRb | myelin protein zero-like 1 | - | HPRD | 15231747 |
| NEK1 | DKFZp686D06121 | DKFZp686K12169 | KIAA1901 | MGC138800 | NY-REN-55 | NIMA (never in mitosis gene a)-related kinase 1 | - | HPRD | 15231747 |
| SKIV2L | 170A | DDX13 | HLP | SKI2 | SKI2W | SKIV2 | superkiller viralicidic activity 2-like (S. cerevisiae) | - | HPRD | 15231747 |
| SMPD4 | FLJ20297 | FLJ20756 | KIAA1418 | NSMASE3 | nSMase-3 | sphingomyelin phosphodiesterase 4, neutral membrane (neutral sphingomyelinase-3) | - | HPRD | 15231747 |
| UPF1 | FLJ43809 | FLJ46894 | HUPF1 | KIAA0221 | NORF1 | RENT1 | pNORF1 | UPF1 regulator of nonsense transcripts homolog (yeast) | - | HPRD,BioGRID | 14527413 |
| UPF2 | DKFZp434D222 | HUPF2 | KIAA1408 | MGC138834 | MGC138835 | RENT2 | smg-3 | UPF2 regulator of nonsense transcripts homolog (yeast) | - | HPRD,BioGRID | 14527413 |
| UPF3A | HUPF3A | RENT3A | UPF3 | UPF3 regulator of nonsense transcripts homolog A (yeast) | Affinity Capture-Western | BioGRID | 14527413 |
| UPF3B | HUPF3B | MRXS14 | RENT3B | UPF3X | UPF3 regulator of nonsense transcripts homolog B (yeast) | - | HPRD | 14527413 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG RNA DEGRADATION | 59 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME DIABETES PATHWAYS | 133 | 91 | All SZGR 2.0 genes in this pathway |
| REACTOME PERK REGULATED GENE EXPRESSION | 29 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF GENES BY ATF4 | 26 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME UNFOLDED PROTEIN RESPONSE | 80 | 51 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF MRNA | 284 | 128 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | 11 | 5 | All SZGR 2.0 genes in this pathway |
| REACTOME DEADENYLATION DEPENDENT MRNA DECAY | 48 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF RNA | 330 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME DESTABILIZATION OF MRNA BY BRF1 | 17 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | 84 | 50 | All SZGR 2.0 genes in this pathway |
| REACTOME DESTABILIZATION OF MRNA BY KSRP | 17 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | 17 | 7 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS DN | 182 | 111 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE DN | 384 | 220 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA UP | 368 | 234 | All SZGR 2.0 genes in this pathway |
| GARGALOVIC RESPONSE TO OXIDIZED PHOSPHOLIPIDS GREY DN | 74 | 44 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 MUTATED SIGNATURE 1 UP | 276 | 165 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 UP | 380 | 236 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 UP | 418 | 263 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 SIGNATURE 3 UP | 341 | 197 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC DN | 187 | 115 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA DN | 146 | 94 | All SZGR 2.0 genes in this pathway |
| CHOW RASSF1 TARGETS DN | 29 | 19 | All SZGR 2.0 genes in this pathway |
| MATTIOLI MGUS VS PCL | 116 | 62 | All SZGR 2.0 genes in this pathway |
| OXFORD RALA OR RALB TARGETS UP | 48 | 23 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN | 637 | 377 | All SZGR 2.0 genes in this pathway |
| CAFFAREL RESPONSE TO THC 24HR 5 UP | 34 | 23 | All SZGR 2.0 genes in this pathway |
| SUH COEXPRESSED WITH ID1 AND ID2 UP | 19 | 14 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 8Q23 Q24 AMPLICON | 157 | 87 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS REQUIRE MYC | 210 | 123 | All SZGR 2.0 genes in this pathway |
| YEGNASUBRAMANIAN PROSTATE CANCER | 128 | 60 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER UP | 298 | 174 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 DN | 374 | 217 | All SZGR 2.0 genes in this pathway |
| LI DCP2 BOUND MRNA | 89 | 57 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |