Gene Page: PPARG
Summary ?
| GeneID | 5468 |
| Symbol | PPARG |
| Synonyms | CIMT1|GLM1|NR1C3|PPARG1|PPARG2|PPARgamma |
| Description | peroxisome proliferator activated receptor gamma |
| Reference | MIM:601487|HGNC:HGNC:9236|Ensembl:ENSG00000132170|HPRD:03288|Vega:OTTHUMG00000129764 |
| Gene type | protein-coding |
| Map location | 3p25 |
| Pascal p-value | 0.006 |
| Sherlock p-value | 0.6 |
| Fetal beta | -0.921 |
| eGene | Cerebellar Hemisphere Cerebellum Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.006 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs633915 | chr1 | 182682157 | PPARG | 5468 | 0.09 | trans | ||
| rs621071 | chr1 | 182682810 | PPARG | 5468 | 0.07 | trans |
Section II. Transcriptome annotation
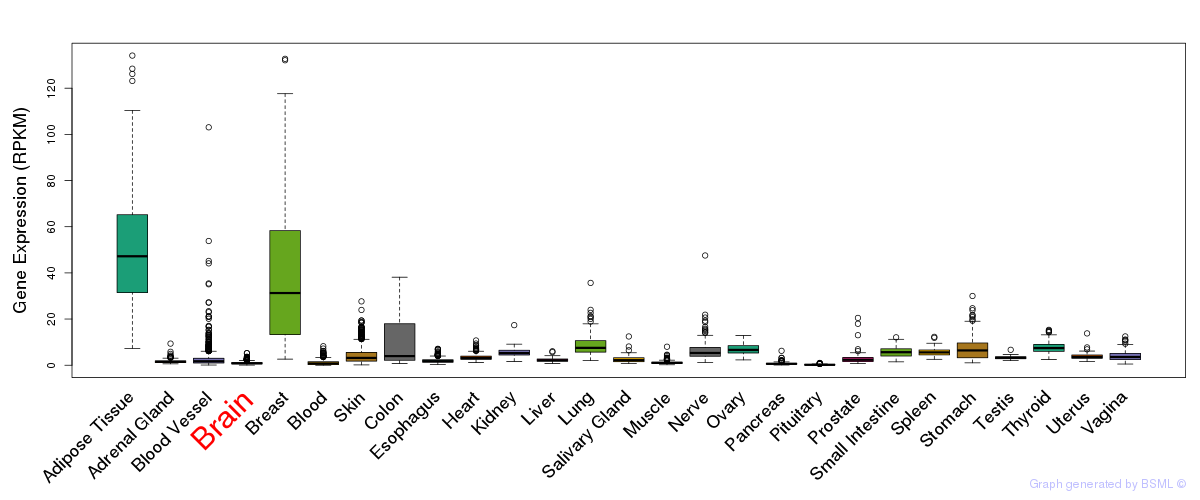
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CMPK1 | 0.94 | 0.94 |
| ATAD1 | 0.93 | 0.93 |
| CNOT7 | 0.93 | 0.93 |
| TTC33 | 0.93 | 0.94 |
| SLC30A9 | 0.92 | 0.92 |
| HSPA13 | 0.92 | 0.93 |
| TBC1D23 | 0.92 | 0.90 |
| STX12 | 0.92 | 0.93 |
| RHOT1 | 0.92 | 0.91 |
| USP33 | 0.92 | 0.91 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FXYD1 | -0.67 | -0.68 |
| MT-CO2 | -0.67 | -0.66 |
| AF347015.33 | -0.64 | -0.64 |
| AF347015.21 | -0.64 | -0.60 |
| AF347015.8 | -0.64 | -0.65 |
| AF347015.2 | -0.63 | -0.62 |
| AC018755.7 | -0.63 | -0.67 |
| MT-CYB | -0.63 | -0.63 |
| HIGD1B | -0.63 | -0.63 |
| AF347015.31 | -0.63 | -0.63 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004872 | receptor activity | IEA | - | |
| GO:0004879 | ligand-dependent nuclear receptor activity | IEA | - | |
| GO:0003700 | transcription factor activity | IEA | - | |
| GO:0003707 | steroid hormone receptor activity | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0004955 | prostaglandin receptor activity | TAS | 9568715 | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0008144 | drug binding | IDA | 9113987 |10622252 | |
| GO:0016564 | transcription repressor activity | IEA | - | |
| GO:0016563 | transcription activator activity | IEA | - | |
| GO:0033613 | transcription activator binding | IDA | 10622252 | |
| GO:0046872 | metal ion binding | IEA | - | |
| GO:0046965 | retinoid X receptor binding | IDA | 9568715 | |
| GO:0046982 | protein heterodimerization activity | TAS | 9744270 | |
| GO:0043565 | sequence-specific DNA binding | IEA | - | |
| GO:0050544 | arachidonic acid binding | ISS | 9568715 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | IEA | - | |
| GO:0001890 | placenta development | ISS | - | |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0030224 | monocyte differentiation | IDA | 9568716 | |
| GO:0042953 | lipoprotein transport | IDA | 9568716 | |
| GO:0006350 | transcription | IEA | - | |
| GO:0008217 | regulation of blood pressure | IMP | 10622252 | |
| GO:0007165 | signal transduction | IDA | 9568716 | |
| GO:0048469 | cell maturation | IDA | 9568716 | |
| GO:0007584 | response to nutrient | TAS | 10973253 | |
| GO:0006629 | lipid metabolic process | TAS | 9568716 | |
| GO:0015909 | long-chain fatty acid transport | ISS | - | |
| GO:0030855 | epithelial cell differentiation | IEA | - | |
| GO:0033993 | response to lipid | ISS | 9568715 | |
| GO:0042593 | glucose homeostasis | IMP | 10622252 | |
| GO:0050872 | white fat cell differentiation | IEA | - | |
| GO:0050872 | white fat cell differentiation | TAS | 12588810 | |
| GO:0032869 | cellular response to insulin stimulus | IMP | 10622252 | |
| GO:0045165 | cell fate commitment | IEA | - | |
| GO:0045600 | positive regulation of fat cell differentiation | IEA | - | |
| GO:0045600 | positive regulation of fat cell differentiation | ISS | - | |
| GO:0045941 | positive regulation of transcription | IEA | - | |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | IEA | - | |
| GO:0045087 | innate immune response | TAS | 17145956 | |
| GO:0045713 | low-density lipoprotein receptor biosynthetic process | IDA | 9568716 | |
| GO:0055088 | lipid homeostasis | TAS | 9113987 | |
| GO:0055098 | response to low density lipoprotein stimulus | IDA | 9568716 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | IEA | - | |
| GO:0005634 | nucleus | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BCAS2 | DAM1 | breast carcinoma amplified sequence 2 | BCAS2 interacts with PPAR-gamma. This interaction was modeled on a demonstrated interaction between human BCAS2 and PPAR-gamma from an unspecified species. | BIND | 15694360 |
| BRD8 | SMAP | SMAP2 | p120 | bromodomain containing 8 | - | HPRD | 10517671 |
| CEP350 | CAP350 | FLJ38282 | FLJ44058 | GM133 | KIAA0480 | centrosomal protein 350kDa | CAP350 interacts with PPAR-gamma-2. | BIND | 15615782 |
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | Reconstituted Complex | BioGRID | 10848596 |
| DNTTIP2 | ERBP | FCF2 | HSU15552 | LPTS-RP2 | MGC163494 | RP4-561L24.1 | TdIF2 | deoxynucleotidyltransferase, terminal, interacting protein 2 | - | HPRD,BioGRID | 1504714 |
| DUT | FLJ20622 | dUTPase | deoxyuridine triphosphatase | - | HPRD | 8910358 |
| EDF1 | EDF-1 | MBF1 | MGC9058 | endothelial differentiation-related factor 1 | - | HPRD,BioGRID | 12040021 |
| EP300 | KAT3B | p300 | E1A binding protein p300 | Reconstituted Complex Two-hybrid | BioGRID | 10944516 |12479814 |
| FABP1 | FABPL | L-FABP | fatty acid binding protein 1, liver | - | HPRD,BioGRID | 11226238 |
| GADD45B | DKFZp566B133 | GADD45BETA | MYD118 | growth arrest and DNA-damage-inducible, beta | - | HPRD | 10872826 |
| GADD45G | CR6 | DDIT2 | GADD45gamma | GRP17 | growth arrest and DNA-damage-inducible, gamma | - | HPRD,BioGRID | 10872826 |
| HDAC3 | HD3 | RPD3 | RPD3-2 | histone deacetylase 3 | - | HPRD,BioGRID | 12479814 |
| HDAC4 | HA6116 | HD4 | HDAC-A | HDACA | KIAA0288 | histone deacetylase 4 | Reconstituted Complex | BioGRID | 12943985 |
| MED1 | CRSP1 | CRSP200 | DRIP205 | DRIP230 | MGC71488 | PBP | PPARBP | PPARGBP | RB18A | TRAP220 | TRIP2 | mediator complex subunit 1 | - | HPRD | 12037571 |
| MED1 | CRSP1 | CRSP200 | DRIP205 | DRIP230 | MGC71488 | PBP | PPARBP | PPARGBP | RB18A | TRAP220 | TRIP2 | mediator complex subunit 1 | Reconstituted Complex Two-hybrid | BioGRID | 10944516 |
| MED24 | ARC100 | CRSP100 | CRSP4 | DRIP100 | KIAA0130 | MGC8748 | THRAP4 | TRAP100 | mediator complex subunit 24 | - | HPRD,BioGRID | 9653119 |
| MPG | AAG | APNG | CRA36.1 | MDG | Mid1 | PIG11 | PIG16 | anpg | N-methylpurine-DNA glycosylase | MPG interacts with an unspecified isoform of PPARG (PPAR-gamma). This interaction was modeled on a demonstrated interaction between MPG from an unspecified species and PPARG from an unspecified species. | BIND | 14761960 |
| NCOA1 | F-SRC-1 | KAT13A | MGC129719 | MGC129720 | NCoA-1 | RIP160 | SRC-1 | SRC1 | bHLHe42 | nuclear receptor coactivator 1 | - | HPRD | 9041124 |10848596 |11330046 |
| NCOA2 | GRIP1 | KAT13C | MGC138808 | NCoA-2 | TIF2 | nuclear receptor coactivator 2 | - | HPRD,BioGRID | 10944516 |
| NCOA3 | ACTR | AIB-1 | AIB1 | CAGH16 | CTG26 | KAT13B | MGC141848 | RAC3 | SRC3 | TNRC14 | TNRC16 | TRAM-1 | pCIP | nuclear receptor coactivator 3 | Reconstituted Complex Two-hybrid | BioGRID | 10944516 |
| NCOA4 | ARA70 | DKFZp762E1112 | ELE1 | PTC3 | RFG | nuclear receptor coactivator 4 | - | HPRD,BioGRID | 10347167 |
| NCOA6 | AIB3 | ANTP | ASC2 | HOX1.1 | HOXA7 | KIAA0181 | NRC | PRIP | RAP250 | TRBP | nuclear receptor coactivator 6 | - | HPRD | 10788465 |
| NCOA6 | AIB3 | ANTP | ASC2 | HOX1.1 | HOXA7 | KIAA0181 | NRC | PRIP | RAP250 | TRBP | nuclear receptor coactivator 6 | Two-hybrid | BioGRID | 11158331 |
| NFE2L2 | NRF2 | nuclear factor (erythroid-derived 2)-like 2 | - | HPRD,BioGRID | 10930400 |
| NR0B2 | FLJ17090 | SHP | SHP1 | nuclear receptor subfamily 0, group B, member 2 | - | HPRD,BioGRID | 11696534 |
| NRIP1 | FLJ77253 | RIP140 | nuclear receptor interacting protein 1 | - | HPRD,BioGRID | 9626662 |
| POU1F1 | GHF-1 | PIT1 | Pit-1 | POU class 1 homeobox 1 | Reconstituted Complex | BioGRID | 11891224 |
| PPARG | NR1C3 | PPARG1 | PPARG2 | PPARgamma | peroxisome proliferator-activated receptor gamma | - | HPRD | 11831892 |
| PPARGC1A | LEM6 | PGC-1(alpha) | PGC-1v | PGC1 | PGC1A | PPARGC1 | peroxisome proliferator-activated receptor gamma, coactivator 1 alpha | - | HPRD | 12502716 |
| PPARGC1A | LEM6 | PGC-1(alpha) | PGC-1v | PGC1 | PGC1A | PPARGC1 | peroxisome proliferator-activated receptor gamma, coactivator 1 alpha | Reconstituted Complex | BioGRID | 14636573 |
| PRMT2 | HRMT1L1 | MGC111373 | protein arginine methyltransferase 2 | PRMT2 interacts with PPAR-gamma. | BIND | 12039952 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 12479814 |
| RXRA | FLJ00280 | FLJ00318 | FLJ16020 | FLJ16733 | MGC102720 | NR2B1 | retinoid X receptor, alpha | - | HPRD,BioGRID | 7838715 |10854698 |
| RXRA | FLJ00280 | FLJ00318 | FLJ16020 | FLJ16733 | MGC102720 | NR2B1 | retinoid X receptor, alpha | - | HPRD | 7838715 |10854698 |10882139 |
| RXRG | NR2B3 | RXRC | retinoid X receptor, gamma | - | HPRD | 12615696 |
| SVIL | DKFZp686A17191 | supervillin | Two-hybrid | BioGRID | 11792840 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PPAR SIGNALING PATHWAY | 69 | 47 | All SZGR 2.0 genes in this pathway |
| KEGG HUNTINGTONS DISEASE | 185 | 109 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG THYROID CANCER | 29 | 26 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NUCLEARRS PATHWAY | 15 | 12 | All SZGR 2.0 genes in this pathway |
| PID WNT NONCANONICAL PATHWAY | 32 | 26 | All SZGR 2.0 genes in this pathway |
| PID NFAT TFPATHWAY | 47 | 39 | All SZGR 2.0 genes in this pathway |
| PID HDAC CLASSI PATHWAY | 66 | 50 | All SZGR 2.0 genes in this pathway |
| PID RXR VDR PATHWAY | 26 | 24 | All SZGR 2.0 genes in this pathway |
| PID RB 1PATHWAY | 65 | 46 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME PPARA ACTIVATES GENE EXPRESSION | 104 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME GENERIC TRANSCRIPTION PATHWAY | 352 | 181 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | 49 | 36 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | 168 | 115 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | 72 | 53 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS UP | 149 | 84 | All SZGR 2.0 genes in this pathway |
| LUI THYROID CANCER CLUSTER 2 | 42 | 31 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| GOTZMANN EPITHELIAL TO MESENCHYMAL TRANSITION DN | 206 | 136 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 UP | 256 | 159 | All SZGR 2.0 genes in this pathway |
| ROZANOV MMP14 TARGETS UP | 266 | 171 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA UP | 207 | 145 | All SZGR 2.0 genes in this pathway |
| LENAOUR DENDRITIC CELL MATURATION UP | 114 | 84 | All SZGR 2.0 genes in this pathway |
| GERY CEBP TARGETS | 126 | 90 | All SZGR 2.0 genes in this pathway |
| KANG IMMORTALIZED BY TERT UP | 89 | 61 | All SZGR 2.0 genes in this pathway |
| PETROVA ENDOTHELIUM LYMPHATIC VS BLOOD UP | 131 | 87 | All SZGR 2.0 genes in this pathway |
| APPEL IMATINIB RESPONSE | 33 | 22 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RESPONSE TO TRABECTEDIN | 50 | 32 | All SZGR 2.0 genes in this pathway |
| URS ADIPOCYTE DIFFERENTIATION UP | 74 | 51 | All SZGR 2.0 genes in this pathway |
| RUAN RESPONSE TO TNF TROGLITAZONE DN | 42 | 24 | All SZGR 2.0 genes in this pathway |
| RUAN RESPONSE TO TNF DN | 84 | 50 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 5 | 126 | 78 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION MUSCLE UP | 43 | 33 | All SZGR 2.0 genes in this pathway |
| CHANG IMMORTALIZED BY HPV31 UP | 84 | 55 | All SZGR 2.0 genes in this pathway |
| LI ADIPOGENESIS BY ACTIVATED PPARG | 17 | 12 | All SZGR 2.0 genes in this pathway |
| GERHOLD ADIPOGENESIS UP | 49 | 40 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL A UP | 84 | 52 | All SZGR 2.0 genes in this pathway |
| BOCHKIS FOXA2 TARGETS | 425 | 261 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| BRUECKNER TARGETS OF MIRLET7A3 DN | 78 | 49 | All SZGR 2.0 genes in this pathway |
| FIRESTEIN PROLIFERATION | 175 | 125 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| PARK TRETINOIN RESPONSE AND RARA PLZF FUSION | 22 | 14 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| LIU VAV3 PROSTATE CARCINOGENESIS UP | 89 | 61 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S2 | 115 | 74 | All SZGR 2.0 genes in this pathway |
| NAKAYAMA SOFT TISSUE TUMORS PCA2 DN | 80 | 51 | All SZGR 2.0 genes in this pathway |
| NAKAMURA ADIPOGENESIS LATE UP | 104 | 67 | All SZGR 2.0 genes in this pathway |
| GERHOLD RESPONSE TO TZD DN | 13 | 11 | All SZGR 2.0 genes in this pathway |
| RUAN RESPONSE TO TROGLITAZONE DN | 19 | 14 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 1HR DN | 222 | 147 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT | 567 | 365 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR DN | 505 | 328 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS NOT VIA AKT1 DN | 88 | 61 | All SZGR 2.0 genes in this pathway |
| STEGER ADIPOGENESIS UP | 21 | 16 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |
| SERVITJA LIVER HNF1A TARGETS UP | 135 | 96 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 4 | 110 | 66 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND WITH H4K20ME1 MARK | 145 | 82 | All SZGR 2.0 genes in this pathway |
| VERNOCHET ADIPOGENESIS | 19 | 11 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE NOT VIA P38 | 337 | 236 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-128 | 82 | 88 | 1A | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-130/301 | 42 | 49 | 1A,m8 | hsa-miR-130abrain | CAGUGCAAUGUUAAAAGGGCAU |
| hsa-miR-301 | CAGUGCAAUAGUAUUGUCAAAGC | ||||
| hsa-miR-130bbrain | CAGUGCAAUGAUGAAAGGGCAU | ||||
| hsa-miR-454-3p | UAGUGCAAUAUUGCUUAUAGGGUUU | ||||
| miR-27 | 82 | 89 | 1A,m8 | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.