Gene Page: ARL8B
Summary ?
| GeneID | 55207 |
| Symbol | ARL8B |
| Synonyms | ARL10C|Gie1 |
| Description | ADP ribosylation factor like GTPase 8B |
| Reference | MIM:616596|HGNC:HGNC:25564|Ensembl:ENSG00000134108|HPRD:12483|Vega:OTTHUMG00000090463 |
| Gene type | protein-coding |
| Map location | 3p26.1 |
| Pascal p-value | 0.108 |
| Sherlock p-value | 0.263 |
| Fetal beta | -1.816 |
| DMG | 1 (# studies) |
| eGene | Caudate basal ganglia Cerebellum Cortex Hippocampus Nucleus accumbens basal ganglia Putamen basal ganglia |
| Support | INTRACELLULAR TRAFFICKING |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 1 |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.066 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg03223878 | 3 | 5021310 | ARL8B | 0.003 | -6.044 | DMG:vanEijk_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs4499578 | 3 | 5167618 | ARL8B | ENSG00000134108.8 | 1.464E-6 | 0.01 | 3713 | gtex_brain_putamen_basal |
| rs62253635 | 3 | 5167997 | ARL8B | ENSG00000134108.8 | 1.464E-6 | 0.01 | 4092 | gtex_brain_putamen_basal |
| rs11926551 | 3 | 5168519 | ARL8B | ENSG00000134108.8 | 1.462E-6 | 0.01 | 4614 | gtex_brain_putamen_basal |
| rs11710829 | 3 | 5170332 | ARL8B | ENSG00000134108.8 | 1.517E-6 | 0.01 | 6427 | gtex_brain_putamen_basal |
| rs57459025 | 3 | 5170865 | ARL8B | ENSG00000134108.8 | 1.464E-6 | 0.01 | 6960 | gtex_brain_putamen_basal |
| rs11915596 | 3 | 5170906 | ARL8B | ENSG00000134108.8 | 1.464E-6 | 0.01 | 7001 | gtex_brain_putamen_basal |
| rs35833834 | 3 | 5171872 | ARL8B | ENSG00000134108.8 | 1.751E-6 | 0.01 | 7967 | gtex_brain_putamen_basal |
| rs200932490 | 3 | 5173073 | ARL8B | ENSG00000134108.8 | 1.419E-6 | 0.01 | 9168 | gtex_brain_putamen_basal |
| rs62253640 | 3 | 5173774 | ARL8B | ENSG00000134108.8 | 1.465E-6 | 0.01 | 9869 | gtex_brain_putamen_basal |
| rs13062913 | 3 | 5173799 | ARL8B | ENSG00000134108.8 | 1.798E-6 | 0.01 | 9894 | gtex_brain_putamen_basal |
| rs13072454 | 3 | 5174223 | ARL8B | ENSG00000134108.8 | 8.797E-7 | 0.01 | 10318 | gtex_brain_putamen_basal |
| rs13072633 | 3 | 5174332 | ARL8B | ENSG00000134108.8 | 1.798E-6 | 0.01 | 10427 | gtex_brain_putamen_basal |
| rs34287764 | 3 | 5174786 | ARL8B | ENSG00000134108.8 | 8.799E-7 | 0.01 | 10881 | gtex_brain_putamen_basal |
| rs35393423 | 3 | 5174908 | ARL8B | ENSG00000134108.8 | 9.195E-7 | 0.01 | 11003 | gtex_brain_putamen_basal |
| rs11719986 | 3 | 5175530 | ARL8B | ENSG00000134108.8 | 8.774E-7 | 0.01 | 11625 | gtex_brain_putamen_basal |
| rs5846363 | 3 | 5178445 | ARL8B | ENSG00000134108.8 | 1.582E-6 | 0.01 | 14540 | gtex_brain_putamen_basal |
| rs11720410 | 3 | 5183861 | ARL8B | ENSG00000134108.8 | 7.975E-7 | 0.01 | 19956 | gtex_brain_putamen_basal |
| rs7633347 | 3 | 5184779 | ARL8B | ENSG00000134108.8 | 7.873E-7 | 0.01 | 20874 | gtex_brain_putamen_basal |
| rs56778477 | 3 | 5186772 | ARL8B | ENSG00000134108.8 | 6.323E-7 | 0.01 | 22867 | gtex_brain_putamen_basal |
| rs55948235 | 3 | 5187031 | ARL8B | ENSG00000134108.8 | 3.789E-7 | 0.01 | 23126 | gtex_brain_putamen_basal |
| rs398091250 | 3 | 5187512 | ARL8B | ENSG00000134108.8 | 8.388E-7 | 0.01 | 23607 | gtex_brain_putamen_basal |
| rs11921280 | 3 | 5188195 | ARL8B | ENSG00000134108.8 | 7.934E-7 | 0.01 | 24290 | gtex_brain_putamen_basal |
| rs11918654 | 3 | 5188246 | ARL8B | ENSG00000134108.8 | 7.934E-7 | 0.01 | 24341 | gtex_brain_putamen_basal |
| rs11929290 | 3 | 5188432 | ARL8B | ENSG00000134108.8 | 7.881E-7 | 0.01 | 24527 | gtex_brain_putamen_basal |
| rs55946647 | 3 | 5188814 | ARL8B | ENSG00000134108.8 | 3.606E-7 | 0.01 | 24909 | gtex_brain_putamen_basal |
| rs11710312 | 3 | 5189234 | ARL8B | ENSG00000134108.8 | 3.656E-7 | 0.01 | 25329 | gtex_brain_putamen_basal |
| rs11710404 | 3 | 5189539 | ARL8B | ENSG00000134108.8 | 3.615E-7 | 0.01 | 25634 | gtex_brain_putamen_basal |
| rs3864060 | 3 | 5189592 | ARL8B | ENSG00000134108.8 | 7.187E-7 | 0.01 | 25687 | gtex_brain_putamen_basal |
| rs11714002 | 3 | 5189701 | ARL8B | ENSG00000134108.8 | 7.544E-7 | 0.01 | 25796 | gtex_brain_putamen_basal |
| rs11706544 | 3 | 5189899 | ARL8B | ENSG00000134108.8 | 7.505E-7 | 0.01 | 25994 | gtex_brain_putamen_basal |
| rs11711365 | 3 | 5189979 | ARL8B | ENSG00000134108.8 | 7.476E-7 | 0.01 | 26074 | gtex_brain_putamen_basal |
| rs57213835 | 3 | 5190000 | ARL8B | ENSG00000134108.8 | 3.585E-7 | 0.01 | 26095 | gtex_brain_putamen_basal |
| rs11707685 | 3 | 5191756 | ARL8B | ENSG00000134108.8 | 2.285E-7 | 0.01 | 27851 | gtex_brain_putamen_basal |
| rs9713188 | 3 | 5192139 | ARL8B | ENSG00000134108.8 | 4.01E-7 | 0.01 | 28234 | gtex_brain_putamen_basal |
| rs3967124 | 3 | 5192194 | ARL8B | ENSG00000134108.8 | 2.023E-7 | 0.01 | 28289 | gtex_brain_putamen_basal |
| rs35108357 | 3 | 5192549 | ARL8B | ENSG00000134108.8 | 2.642E-7 | 0.01 | 28644 | gtex_brain_putamen_basal |
| rs200429478 | 3 | 5192551 | ARL8B | ENSG00000134108.8 | 2.792E-7 | 0.01 | 28646 | gtex_brain_putamen_basal |
| rs4054852 | 3 | 5192552 | ARL8B | ENSG00000134108.8 | 4.087E-7 | 0.01 | 28647 | gtex_brain_putamen_basal |
| rs4266178 | 3 | 5192553 | ARL8B | ENSG00000134108.8 | 7.764E-7 | 0.01 | 28648 | gtex_brain_putamen_basal |
| rs12497542 | 3 | 5193541 | ARL8B | ENSG00000134108.8 | 9.177E-7 | 0.01 | 29636 | gtex_brain_putamen_basal |
| rs60416950 | 3 | 5194437 | ARL8B | ENSG00000134108.8 | 7.873E-7 | 0.01 | 30532 | gtex_brain_putamen_basal |
| rs10428135 | 3 | 5195080 | ARL8B | ENSG00000134108.8 | 1.638E-7 | 0.01 | 31175 | gtex_brain_putamen_basal |
| rs10428151 | 3 | 5195655 | ARL8B | ENSG00000134108.8 | 1.55E-7 | 0.01 | 31750 | gtex_brain_putamen_basal |
| rs3899068 | 3 | 5196050 | ARL8B | ENSG00000134108.8 | 1.563E-7 | 0.01 | 32145 | gtex_brain_putamen_basal |
| rs34477772 | 3 | 5196058 | ARL8B | ENSG00000134108.8 | 3.43E-7 | 0.01 | 32153 | gtex_brain_putamen_basal |
| rs3967123 | 3 | 5196406 | ARL8B | ENSG00000134108.8 | 1.554E-7 | 0.01 | 32501 | gtex_brain_putamen_basal |
| rs4054850 | 3 | 5196508 | ARL8B | ENSG00000134108.8 | 1.549E-7 | 0.01 | 32603 | gtex_brain_putamen_basal |
| rs6798707 | 3 | 5196639 | ARL8B | ENSG00000134108.8 | 1.421E-7 | 0.01 | 32734 | gtex_brain_putamen_basal |
| rs6798806 | 3 | 5196707 | ARL8B | ENSG00000134108.8 | 3.43E-7 | 0.01 | 32802 | gtex_brain_putamen_basal |
| rs56215613 | 3 | 5197318 | ARL8B | ENSG00000134108.8 | 2.257E-7 | 0.01 | 33413 | gtex_brain_putamen_basal |
| rs13068579 | 3 | 5198524 | ARL8B | ENSG00000134108.8 | 2.004E-7 | 0.01 | 34619 | gtex_brain_putamen_basal |
| rs11927499 | 3 | 5199198 | ARL8B | ENSG00000134108.8 | 3.476E-7 | 0.01 | 35293 | gtex_brain_putamen_basal |
| rs62255880 | 3 | 5199351 | ARL8B | ENSG00000134108.8 | 1.405E-6 | 0.01 | 35446 | gtex_brain_putamen_basal |
| rs60724392 | 3 | 5199858 | ARL8B | ENSG00000134108.8 | 1.387E-6 | 0.01 | 35953 | gtex_brain_putamen_basal |
| rs6787725 | 3 | 5201054 | ARL8B | ENSG00000134108.8 | 1.435E-7 | 0.01 | 37149 | gtex_brain_putamen_basal |
| rs13093192 | 3 | 5201615 | ARL8B | ENSG00000134108.8 | 2.004E-7 | 0.01 | 37710 | gtex_brain_putamen_basal |
| rs13093380 | 3 | 5201723 | ARL8B | ENSG00000134108.8 | 2.004E-7 | 0.01 | 37818 | gtex_brain_putamen_basal |
| rs11708037 | 3 | 5201815 | ARL8B | ENSG00000134108.8 | 1.547E-7 | 0.01 | 37910 | gtex_brain_putamen_basal |
| rs60842168 | 3 | 5202445 | ARL8B | ENSG00000134108.8 | 3.426E-7 | 0.01 | 38540 | gtex_brain_putamen_basal |
| rs7614778 | 3 | 5202793 | ARL8B | ENSG00000134108.8 | 1.989E-7 | 0.01 | 38888 | gtex_brain_putamen_basal |
| rs73807499 | 3 | 5202992 | ARL8B | ENSG00000134108.8 | 1.547E-7 | 0.01 | 39087 | gtex_brain_putamen_basal |
| rs11130291 | 3 | 5203776 | ARL8B | ENSG00000134108.8 | 9.86E-8 | 0.01 | 39871 | gtex_brain_putamen_basal |
| rs6803009 | 3 | 5210178 | ARL8B | ENSG00000134108.8 | 1.991E-7 | 0.01 | 46273 | gtex_brain_putamen_basal |
| rs59478365 | 3 | 5211256 | ARL8B | ENSG00000134108.8 | 5.183E-7 | 0.01 | 47351 | gtex_brain_putamen_basal |
| rs6764910 | 3 | 5211390 | ARL8B | ENSG00000134108.8 | 2.004E-7 | 0.01 | 47485 | gtex_brain_putamen_basal |
| rs3856889 | 3 | 5212903 | ARL8B | ENSG00000134108.8 | 9.86E-8 | 0.01 | 48998 | gtex_brain_putamen_basal |
| rs2270750 | 3 | 5214126 | ARL8B | ENSG00000134108.8 | 3.503E-7 | 0.01 | 50221 | gtex_brain_putamen_basal |
| rs7614298 | 3 | 5219198 | ARL8B | ENSG00000134108.8 | 1.373E-7 | 0.01 | 55293 | gtex_brain_putamen_basal |
| rs59759123 | 3 | 5222488 | ARL8B | ENSG00000134108.8 | 3.456E-7 | 0.01 | 58583 | gtex_brain_putamen_basal |
| rs455832 | 3 | 5225783 | ARL8B | ENSG00000134108.8 | 5.558E-7 | 0.01 | 61878 | gtex_brain_putamen_basal |
| rs6765898 | 3 | 5227663 | ARL8B | ENSG00000134108.8 | 9.41E-7 | 0.01 | 63758 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
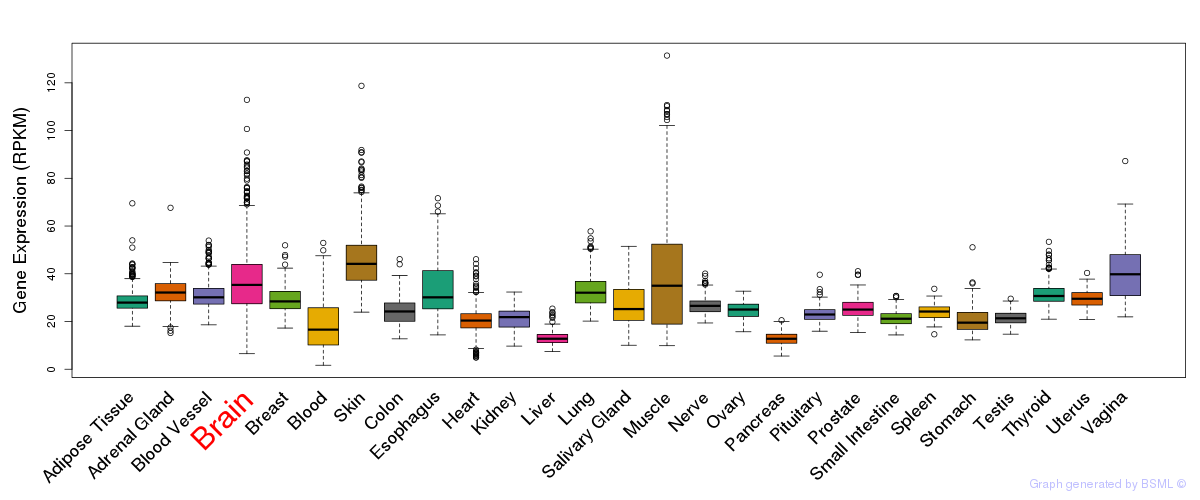
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0003924 | GTPase activity | NAS | 15331635 | |
| GO:0005525 | GTP binding | IDA | 15331635 | |
| GO:0019003 | GDP binding | IDA | 15331635 | |
| GO:0043014 | alpha-tubulin binding | IDA | 15331635 | |
| GO:0048487 | beta-tubulin binding | IDA | 15331635 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007264 | small GTPase mediated signal transduction | IEA | - | |
| GO:0007059 | chromosome segregation | IMP | 14871887 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005737 | cytoplasm | IDA | 15331635 | |
| GO:0005765 | lysosomal membrane | IEA | - | |
| GO:0005768 | endosome | IEA | - | |
| GO:0016020 | membrane | IEA | - | |
| GO:0030496 | midbody | IDA | 15331635 | |
| GO:0051233 | spindle midzone | IDA | 15331635 | |
| GO:0031902 | late endosome membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE UP | 423 | 283 | All SZGR 2.0 genes in this pathway |
| LAIHO COLORECTAL CANCER SERRATED UP | 112 | 71 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER DN | 514 | 319 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY DN | 382 | 224 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED MODERATELY VS POORLY UP | 121 | 71 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA CANCER | 83 | 52 | All SZGR 2.0 genes in this pathway |
| JEPSEN SMRT TARGETS | 33 | 23 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN | 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN | 442 | 275 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 7 | 76 | 46 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 DN | 448 | 282 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-103/107 | 917 | 923 | 1A | hsa-miR-103brain | AGCAGCAUUGUACAGGGCUAUGA |
| hsa-miR-107brain | AGCAGCAUUGUACAGGGCUAUCA | ||||
| miR-124.1 | 1492 | 1498 | 1A | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-128 | 1358 | 1364 | m8 | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-148/152 | 133 | 139 | m8 | hsa-miR-148a | UCAGUGCACUACAGAACUUUGU |
| hsa-miR-152brain | UCAGUGCAUGACAGAACUUGGG | ||||
| hsa-miR-148b | UCAGUGCAUCACAGAACUUUGU | ||||
| miR-15/16/195/424/497 | 914 | 920 | m8 | hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG |
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| miR-19 | 1528 | 1534 | 1A | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-200bc/429 | 450 | 456 | m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-204/211 | 1259 | 1265 | m8 | hsa-miR-204brain | UUCCCUUUGUCAUCCUAUGCCU |
| hsa-miR-211 | UUCCCUUUGUCAUCCUUCGCCU | ||||
| miR-214 | 137 | 143 | 1A | hsa-miR-214brain | ACAGCAGGCACAGACAGGCAG |
| miR-218 | 974 | 980 | 1A | hsa-miR-218brain | UUGUGCUUGAUCUAACCAUGU |
| miR-23 | 1909 | 1915 | 1A | hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC |
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC | ||||
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| miR-30-5p | 1984 | 1990 | m8 | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-320 | 2000 | 2007 | 1A,m8 | hsa-miR-320 | AAAAGCUGGGUUGAGAGGGCGAA |
| miR-34/449 | 1483 | 1489 | 1A | hsa-miR-34abrain | UGGCAGUGUCUUAGCUGGUUGUU |
| hsa-miR-34c | AGGCAGUGUAGUUAGCUGAUUGC | ||||
| hsa-miR-449 | UGGCAGUGUAUUGUUAGCUGGU | ||||
| hsa-miR-449b | AGGCAGUGUAUUGUUAGCUGGC | ||||
| miR-377 | 1363 | 1369 | m8 | hsa-miR-377 | AUCACACAAAGGCAACUUUUGU |
| miR-433-3p | 2062 | 2068 | m8 | hsa-miR-433brain | AUCAUGAUGGGCUCCUCGGUGU |
| miR-539 | 123 | 130 | 1A,m8 | hsa-miR-539 | GGAGAAAUUAUCCUUGGUGUGU |
| miR-7 | 14 | 21 | 1A,m8 | hsa-miR-7SZ | UGGAAGACUAGUGAUUUUGUUG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.