Gene Page: PRKAB1
Summary ?
| GeneID | 5564 |
| Symbol | PRKAB1 |
| Synonyms | AMPK|HAMPKb |
| Description | protein kinase AMP-activated non-catalytic subunit beta 1 |
| Reference | MIM:602740|HGNC:HGNC:9378|Ensembl:ENSG00000111725|HPRD:04116|Vega:OTTHUMG00000168954 |
| Gene type | protein-coding |
| Map location | 12q24.1-q24.3 |
| Pascal p-value | 0.001 |
| Fetal beta | 0.41 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.126 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs13291971 | chr9 | 12861064 | PRKAB1 | 5564 | 0.16 | trans |
Section II. Transcriptome annotation
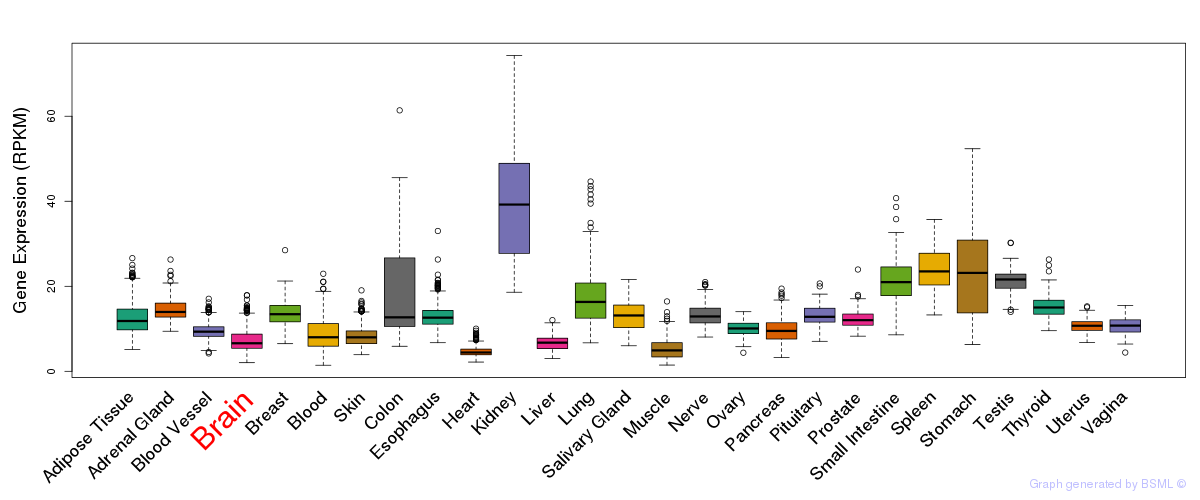
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PDE7B | 0.80 | 0.78 |
| INF2 | 0.79 | 0.74 |
| FBXL16 | 0.78 | 0.77 |
| IL17RA | 0.77 | 0.77 |
| SMAD3 | 0.76 | 0.79 |
| SLC6A1 | 0.75 | 0.85 |
| STIM1 | 0.75 | 0.77 |
| AGFG2 | 0.75 | 0.75 |
| ACTN1 | 0.74 | 0.71 |
| RAP1GAP | 0.73 | 0.66 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RPL24 | -0.48 | -0.59 |
| RPS3AP47 | -0.47 | -0.55 |
| RPL31 | -0.46 | -0.57 |
| RPL27 | -0.46 | -0.56 |
| RPL35A | -0.46 | -0.61 |
| C12orf45 | -0.46 | -0.54 |
| RPS23 | -0.46 | -0.57 |
| RPS20 | -0.46 | -0.61 |
| EXOSC8 | -0.46 | -0.50 |
| C9orf46 | -0.45 | -0.54 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 18403135 | |
| GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005975 | carbohydrate metabolic process | IEA | - | |
| GO:0007165 | signal transduction | TAS | 9575201 | |
| GO:0006633 | fatty acid biosynthetic process | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005737 | cytoplasm | IDA | 18029348 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACLY | ACL | ATPCL | CLATP | ATP citrate lyase | Affinity Capture-MS | BioGRID | 17353931 |
| DHX36 | DDX36 | G4R1 | KIAA1488 | MLEL1 | RHAU | DEAH (Asp-Glu-Ala-His) box polypeptide 36 | Affinity Capture-MS | BioGRID | 17353931 |
| EZR | CVIL | CVL | DKFZp762H157 | FLJ26216 | MGC1584 | VIL2 | ezrin | Affinity Capture-MS | BioGRID | 17353931 |
| F10 | FX | FXA | coagulation factor X | Two-hybrid | BioGRID | 16169070 |
| FNIP1 | DKFZp686E18167 | DKFZp781P0215 | KIAA1961 | MGC667 | folliculin interacting protein 1 | Affinity Capture-Western | BioGRID | 17028174 |
| GYS1 | GSY | GYS | glycogen synthase 1 (muscle) | Affinity Capture-MS | BioGRID | 17353931 |
| MARS | FLJ35667 | METRS | MTRNS | methionyl-tRNA synthetase | Affinity Capture-MS | BioGRID | 17353931 |
| MYH10 | MGC134913 | MGC134914 | NMMHCB | myosin, heavy chain 10, non-muscle | Affinity Capture-MS | BioGRID | 17353931 |
| NDUFA7 | B14.5a | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kDa | Two-hybrid | BioGRID | 16169070 |
| PRKAB2 | MGC61468 | protein kinase, AMP-activated, beta 2 non-catalytic subunit | Affinity Capture-MS | BioGRID | 17353931 |
| PRKAG1 | AMPKG | MGC8666 | protein kinase, AMP-activated, gamma 1 non-catalytic subunit | - | HPRD,BioGRID | 10698692 |
| PRKAG2 | AAKG | AAKG2 | CMH6 | H91620p | WPWS | protein kinase, AMP-activated, gamma 2 non-catalytic subunit | - | HPRD,BioGRID | 10698692 |
| SNRNP200 | ASCC3L1 | BRR2 | HELIC2 | U5-200KD | small nuclear ribonucleoprotein 200kDa (U5) | Affinity Capture-MS | BioGRID | 17353931 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG INSULIN SIGNALING PATHWAY | 137 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG ADIPOCYTOKINE SIGNALING PATHWAY | 67 | 57 | All SZGR 2.0 genes in this pathway |
| KEGG HYPERTROPHIC CARDIOMYOPATHY HCM | 85 | 65 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CHREBP2 PATHWAY | 42 | 35 | All SZGR 2.0 genes in this pathway |
| BIOCARTA LEPTIN PATHWAY | 11 | 11 | All SZGR 2.0 genes in this pathway |
| PID LKB1 PATHWAY | 47 | 37 | All SZGR 2.0 genes in this pathway |
| PID P53 DOWNSTREAM PATHWAY | 137 | 94 | All SZGR 2.0 genes in this pathway |
| PID VEGFR1 2 PATHWAY | 69 | 57 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | 19 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME INSULIN RECEPTOR SIGNALLING CASCADE | 87 | 64 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | 15 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME ENERGY DEPENDENT REGULATION OF MTOR BY LKB1 AMPK | 18 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | 10 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME PKB MEDIATED EVENTS | 29 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY INSULIN RECEPTOR | 108 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME PI3K CASCADE | 71 | 51 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS DN | 384 | 230 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 MUTATED SIGNATURE 2 UP | 139 | 83 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 SIGNATURE 3 UP | 341 | 197 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS UP | 293 | 179 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| KANNAN TP53 TARGETS UP | 58 | 40 | All SZGR 2.0 genes in this pathway |
| WESTON VEGFA TARGETS 6HR | 59 | 38 | All SZGR 2.0 genes in this pathway |
| WESTON VEGFA TARGETS | 108 | 71 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY 4NQO OR UV | 63 | 44 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE DN | 195 | 135 | All SZGR 2.0 genes in this pathway |
| AMBROSINI FLAVOPIRIDOL TREATMENT TP53 | 109 | 63 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| NICK RESPONSE TO PROC TREATMENT DN | 27 | 18 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS UNANNOTATED DN | 193 | 112 | All SZGR 2.0 genes in this pathway |
| KYNG WERNER SYNDROM AND NORMAL AGING UP | 93 | 62 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 9 | 76 | 45 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 4HR DN | 254 | 158 | All SZGR 2.0 genes in this pathway |
| SMIRNOV RESPONSE TO IR 6HR UP | 166 | 97 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-137 | 704 | 710 | 1A | hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG |
| miR-539 | 748 | 754 | m8 | hsa-miR-539 | GGAGAAAUUAUCCUUGGUGUGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.