Gene Page: PKIB
Summary ?
| GeneID | 5570 |
| Symbol | PKIB |
| Synonyms | PRKACN2 |
| Description | protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| Reference | MIM:606914|HGNC:HGNC:9018|Ensembl:ENSG00000135549|HPRD:06075|Vega:OTTHUMG00000015488 |
| Gene type | protein-coding |
| Map location | 6q22.31 |
| Pascal p-value | 0.024 |
| Fetal beta | -2.387 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg03902233 | 6 | 122792987 | PKIB;SERINC1 | 1.358E-4 | -0.338 | 0.031 | DMG:Wockner_2014 |
| cg20242210 | 6 | 122931702 | PKIB | 2.816E-4 | -0.196 | 0.039 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1954436 | chr1 | 62248194 | PKIB | 5570 | 0.14 | trans | ||
| rs13447530 | chr1 | 91983798 | PKIB | 5570 | 0.09 | trans | ||
| rs17022378 | chr1 | 206654057 | PKIB | 5570 | 0.05 | trans | ||
| rs6734526 | chr2 | 5616370 | PKIB | 5570 | 0.16 | trans | ||
| rs6753473 | chr2 | 26526418 | PKIB | 5570 | 0.03 | trans | ||
| rs17028363 | chr2 | 41913161 | PKIB | 5570 | 0 | trans | ||
| rs16829545 | chr2 | 151977407 | PKIB | 5570 | 1.061E-40 | trans | ||
| rs16841750 | chr2 | 158288461 | PKIB | 5570 | 2.226E-4 | trans | ||
| rs16846743 | chr2 | 163321013 | PKIB | 5570 | 0.17 | trans | ||
| rs16863816 | chr2 | 177292762 | PKIB | 5570 | 0.03 | trans | ||
| rs7584986 | chr2 | 184111432 | PKIB | 5570 | 3.741E-6 | trans | ||
| rs6741060 | chr2 | 217169088 | PKIB | 5570 | 0.01 | trans | ||
| rs10183246 | chr2 | 237187882 | PKIB | 5570 | 0.05 | trans | ||
| rs10195546 | chr2 | 237187933 | PKIB | 5570 | 0.15 | trans | ||
| rs9810143 | chr3 | 5060209 | PKIB | 5570 | 2.828E-7 | trans | ||
| rs6797307 | chr3 | 8601563 | PKIB | 5570 | 0 | trans | ||
| rs4831224 | chr3 | 115754105 | PKIB | 5570 | 0.14 | trans | ||
| rs6773248 | chr3 | 174790333 | PKIB | 5570 | 0.06 | trans | ||
| rs6797728 | chr3 | 174790417 | PKIB | 5570 | 0.06 | trans | ||
| rs12642340 | chr4 | 35390674 | PKIB | 5570 | 0.15 | trans | ||
| snp_a-1815771 | 0 | PKIB | 5570 | 0.02 | trans | |||
| rs7654426 | chr4 | 86236933 | PKIB | 5570 | 0.2 | trans | ||
| rs17008119 | chr4 | 125321425 | PKIB | 5570 | 0.02 | trans | ||
| rs7692715 | chr4 | 125335209 | PKIB | 5570 | 0 | trans | ||
| rs2183142 | chr4 | 159232695 | PKIB | 5570 | 0.09 | trans | ||
| rs170776 | chr4 | 173276735 | PKIB | 5570 | 0.05 | trans | ||
| rs1396222 | chr4 | 173279496 | PKIB | 5570 | 0.03 | trans | ||
| rs335993 | chr4 | 173321789 | PKIB | 5570 | 0.09 | trans | ||
| rs335980 | chr4 | 173329784 | PKIB | 5570 | 0.02 | trans | ||
| rs335982 | chr4 | 173330945 | PKIB | 5570 | 0.08 | trans | ||
| rs337984 | chr4 | 173411662 | PKIB | 5570 | 0.18 | trans | ||
| rs4488887 | chr4 | 177695833 | PKIB | 5570 | 0.08 | trans | ||
| rs10491487 | chr5 | 80323367 | PKIB | 5570 | 0.08 | trans | ||
| rs10474151 | chr5 | 84286402 | PKIB | 5570 | 0 | trans | ||
| rs13360496 | 0 | PKIB | 5570 | 0.01 | trans | |||
| rs13191953 | chr6 | 24151042 | PKIB | 5570 | 0.01 | trans | ||
| rs3789226 | chr6 | 24286501 | PKIB | 5570 | 0.1 | trans | ||
| rs6907864 | chr6 | 24293323 | PKIB | 5570 | 0.17 | trans | ||
| rs12196880 | chr6 | 24313746 | PKIB | 5570 | 0 | trans | ||
| rs10806988 | chr6 | 24341768 | PKIB | 5570 | 0 | trans | ||
| rs16890367 | chr6 | 38078448 | PKIB | 5570 | 9.85E-5 | trans | ||
| rs12706918 | chr7 | 80266147 | PKIB | 5570 | 0.05 | trans | ||
| rs7787830 | chr7 | 98797019 | PKIB | 5570 | 0.17 | trans | ||
| rs6996695 | chr8 | 77540580 | PKIB | 5570 | 1.254E-4 | trans | ||
| rs3118341 | chr9 | 25185518 | PKIB | 5570 | 0 | trans | ||
| rs1126130 | chr9 | 25209346 | PKIB | 5570 | 0.06 | trans | ||
| rs1998746 | chr9 | 25211761 | PKIB | 5570 | 0.06 | trans | ||
| rs9406868 | chr9 | 25223372 | PKIB | 5570 | 9.538E-4 | trans | ||
| rs11139334 | chr9 | 84209393 | PKIB | 5570 | 1.407E-4 | trans | ||
| rs2393316 | chr10 | 59333070 | PKIB | 5570 | 0.03 | trans | ||
| rs17124813 | chr10 | 110347464 | PKIB | 5570 | 0.08 | trans | ||
| rs11061703 | chr12 | 1436977 | PKIB | 5570 | 0.09 | trans | ||
| rs7972699 | chr12 | 105155290 | PKIB | 5570 | 0.06 | trans | ||
| rs7488439 | chr12 | 105155814 | PKIB | 5570 | 0.02 | trans | ||
| rs17036489 | chr12 | 105292283 | PKIB | 5570 | 0.01 | trans | ||
| rs7337967 | chr13 | 53519044 | PKIB | 5570 | 0.02 | trans | ||
| rs1530404 | chr13 | 64207519 | PKIB | 5570 | 0.1 | trans | ||
| rs17289777 | chr13 | 64223062 | PKIB | 5570 | 0.1 | trans | ||
| rs9989228 | chr14 | 37809252 | PKIB | 5570 | 0.04 | trans | ||
| rs16955618 | chr15 | 29937543 | PKIB | 5570 | 1.22E-36 | trans | ||
| rs2077735 | chr15 | 58479024 | PKIB | 5570 | 0.02 | trans | ||
| rs9941296 | chr16 | 2759498 | PKIB | 5570 | 0.05 | trans | ||
| rs16945437 | chr17 | 11797650 | PKIB | 5570 | 0.14 | trans | ||
| rs4795850 | chr17 | 32383815 | PKIB | 5570 | 0.18 | trans | ||
| rs4795851 | chr17 | 32384050 | PKIB | 5570 | 0.15 | trans | ||
| rs1003837 | chr17 | 32453154 | PKIB | 5570 | 0.16 | trans | ||
| rs11873184 | chr18 | 1584081 | PKIB | 5570 | 2.111E-4 | trans | ||
| rs17681878 | chr18 | 34767223 | PKIB | 5570 | 0.07 | trans | ||
| rs7274477 | chr20 | 1676942 | PKIB | 5570 | 0.01 | trans | ||
| rs1041786 | chr21 | 22617710 | PKIB | 5570 | 2.129E-7 | trans | ||
| rs16986332 | chrX | 10958354 | PKIB | 5570 | 0.15 | trans | ||
| rs16990575 | chrX | 32691327 | PKIB | 5570 | 1.775E-4 | trans | ||
| rs16993189 | chrX | 144666166 | PKIB | 5570 | 0.02 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
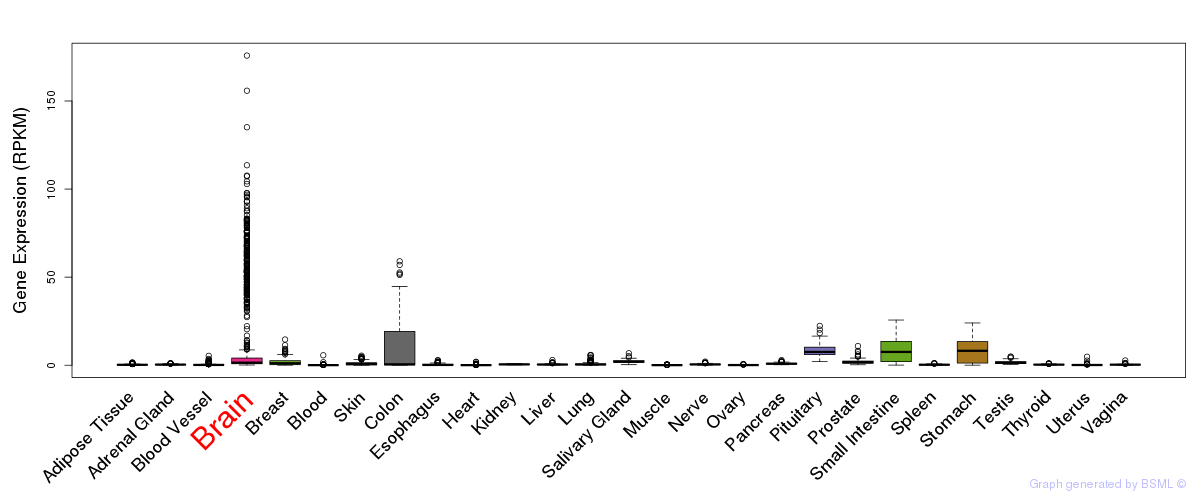
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MAPK9 | 0.92 | 0.89 |
| IQWD1 | 0.91 | 0.90 |
| NAPB | 0.91 | 0.90 |
| ATP6V1C1 | 0.90 | 0.89 |
| IDH3A | 0.90 | 0.90 |
| GABRA1 | 0.90 | 0.92 |
| NSF | 0.90 | 0.88 |
| GLRB | 0.90 | 0.86 |
| PAK1 | 0.90 | 0.85 |
| OXR1 | 0.89 | 0.91 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| BCL7C | -0.46 | -0.54 |
| HYAL2 | -0.42 | -0.42 |
| SH2D2A | -0.42 | -0.45 |
| SH3BP2 | -0.41 | -0.44 |
| ANP32C | -0.41 | -0.41 |
| NME4 | -0.41 | -0.53 |
| SLC38A5 | -0.40 | -0.45 |
| CLEC3B | -0.39 | -0.46 |
| AF347015.18 | -0.39 | -0.18 |
| FADS2 | -0.39 | -0.32 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004862 | cAMP-dependent protein kinase inhibitor activity | IEA | - | |
| GO:0004860 | protein kinase inhibitor activity | IEA | - | |
| GO:0016301 | kinase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006469 | negative regulation of protein kinase activity | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| SENGUPTA NASOPHARYNGEAL CARCINOMA WITH LMP1 UP | 408 | 247 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS UP | 579 | 346 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY DN | 367 | 220 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 16D UP | 175 | 108 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 AND HDAC2 TARGETS DN | 232 | 139 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC2 TARGETS DN | 133 | 77 | All SZGR 2.0 genes in this pathway |
| SABATES COLORECTAL ADENOMA DN | 291 | 176 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 UP | 329 | 196 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM5 | 94 | 59 | All SZGR 2.0 genes in this pathway |
| WILLIAMS ESR1 TARGETS UP | 26 | 16 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| LIAO METASTASIS | 539 | 324 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION DN | 517 | 309 | All SZGR 2.0 genes in this pathway |
| UEDA CENTRAL CLOCK | 88 | 62 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY NO BLOOD UP | 222 | 139 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 4 | 307 | 185 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE DN | 258 | 160 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN UP | 181 | 112 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| HOFFMANN PRE BI TO LARGE PRE BII LYMPHOCYTE DN | 75 | 61 | All SZGR 2.0 genes in this pathway |
| HOFFMANN LARGE TO SMALL PRE BII LYMPHOCYTE DN | 72 | 47 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3 UNMETHYLATED | 536 | 296 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT | 567 | 365 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 6HR UP | 229 | 149 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR UP | 324 | 193 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS NOT VIA AKT1 UP | 211 | 131 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS VIA AKT1 UP | 281 | 183 | All SZGR 2.0 genes in this pathway |