Gene Page: PRL
Summary ?
| GeneID | 5617 |
| Symbol | PRL |
| Synonyms | GHA1 |
| Description | prolactin |
| Reference | MIM:176760|HGNC:HGNC:9445|Ensembl:ENSG00000172179|HPRD:07180|Vega:OTTHUMG00000016111 |
| Gene type | protein-coding |
| Map location | 6p22.3 |
| Pascal p-value | 0.703 |
| Fetal beta | 0.32 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.033 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
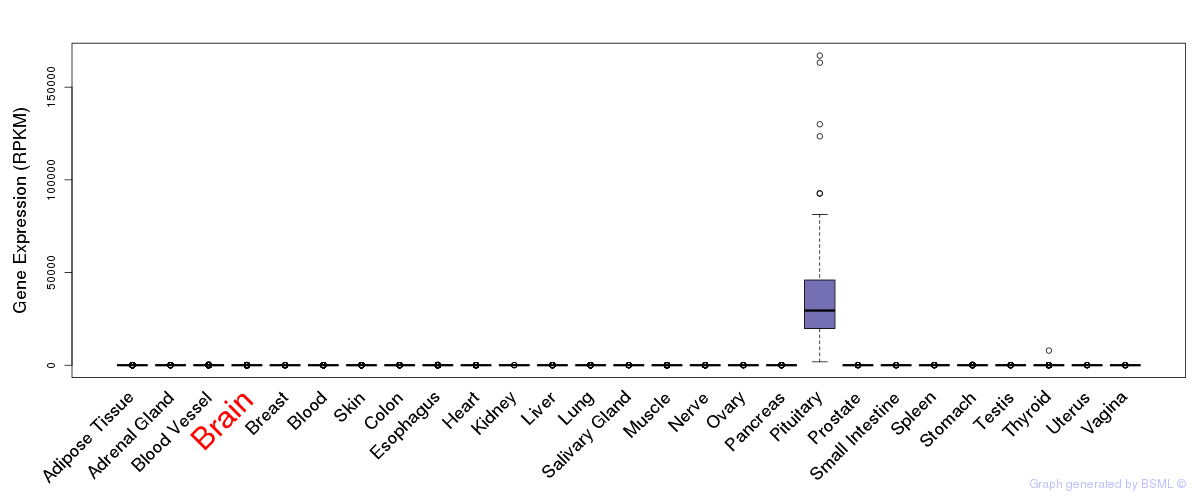
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RYBP | 0.75 | 0.79 |
| PCDHB12 | 0.73 | 0.78 |
| RBM9 | 0.73 | 0.76 |
| CSRNP2 | 0.73 | 0.79 |
| SEC24D | 0.72 | 0.75 |
| EBF3 | 0.72 | 0.62 |
| PCDHB13 | 0.72 | 0.76 |
| IGFBP5 | 0.72 | 0.77 |
| C4orf31 | 0.72 | 0.78 |
| AC093310.1 | 0.72 | 0.77 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.52 | -0.76 |
| AF347015.31 | -0.51 | -0.73 |
| AF347015.33 | -0.51 | -0.72 |
| AF347015.27 | -0.51 | -0.71 |
| SERPINB6 | -0.50 | -0.60 |
| FXYD1 | -0.50 | -0.69 |
| MT-CYB | -0.50 | -0.71 |
| S100B | -0.50 | -0.66 |
| C5orf53 | -0.49 | -0.57 |
| HLA-F | -0.49 | -0.58 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005148 | prolactin receptor binding | TAS | 10854700 | |
| GO:0005179 | hormone activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007166 | cell surface receptor linked signal transduction | TAS | 10854700 | |
| GO:0008283 | cell proliferation | TAS | 10854700 | |
| GO:0007595 | lactation | IEA | - | |
| GO:0007565 | female pregnancy | NAS | 7221563 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CYTOKINE CYTOKINE RECEPTOR INTERACTION | 267 | 161 | All SZGR 2.0 genes in this pathway |
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| KEGG JAK STAT SIGNALING PATHWAY | 155 | 105 | All SZGR 2.0 genes in this pathway |
| PID ERBB4 PATHWAY | 38 | 32 | All SZGR 2.0 genes in this pathway |
| PID PTP1B PATHWAY | 52 | 40 | All SZGR 2.0 genes in this pathway |
| PID REG GR PATHWAY | 82 | 60 | All SZGR 2.0 genes in this pathway |
| PID ERA GENOMIC PATHWAY | 65 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME GROWTH HORMONE RECEPTOR SIGNALING | 24 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB4 | 90 | 67 | All SZGR 2.0 genes in this pathway |
| REACTOME PROLACTIN RECEPTOR SIGNALING | 14 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEAR SIGNALING BY ERBB4 | 38 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM | 270 | 204 | All SZGR 2.0 genes in this pathway |
| REACTOME AMYLOIDS | 83 | 63 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT UP | 390 | 242 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN MGMT 48HR DN | 161 | 105 | All SZGR 2.0 genes in this pathway |
| WANG LSD1 TARGETS DN | 39 | 30 | All SZGR 2.0 genes in this pathway |
| MATZUK LUTEAL GENES | 8 | 5 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS UP | 165 | 118 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER LATE RECURRENCE DN | 69 | 48 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP D | 280 | 158 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 DN | 448 | 282 | All SZGR 2.0 genes in this pathway |
| NABA SECRETED FACTORS | 344 | 197 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |