Gene Page: MARK4
Summary ?
| GeneID | 57787 |
| Symbol | MARK4 |
| Synonyms | MARK4L|MARK4S|MARKL1|MARKL1L|PAR-1D |
| Description | microtubule affinity regulating kinase 4 |
| Reference | MIM:606495|HGNC:HGNC:13538|Ensembl:ENSG00000007047|HPRD:09402|Vega:OTTHUMG00000181769 |
| Gene type | protein-coding |
| Map location | 19q13.3 |
| Pascal p-value | 0.183 |
| TADA p-value | 0.002 |
| Fetal beta | -0.507 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Montano_2016 | Genome-wide DNA methylation analysis | This dataset includes 172 replicated associations between CpGs with schizophrenia. | 1 |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 6 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| MARK4 | chr19 | 45800933 | G | A | NM_001199867 NM_001199867 NM_031417 NM_031417 | . . . . | intronic splice-acceptor-in2 intronic splice-acceptor-in2 | Schizophrenia | DNM:Fromer_2014 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg14114377 | 19 | 45756845 | MARK4 | 1.07E-5 | -0.008 | 0.045 | DMG:Montano_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10994209 | chr10 | 61877705 | MARK4 | 57787 | 0.19 | trans |
Section II. Transcriptome annotation
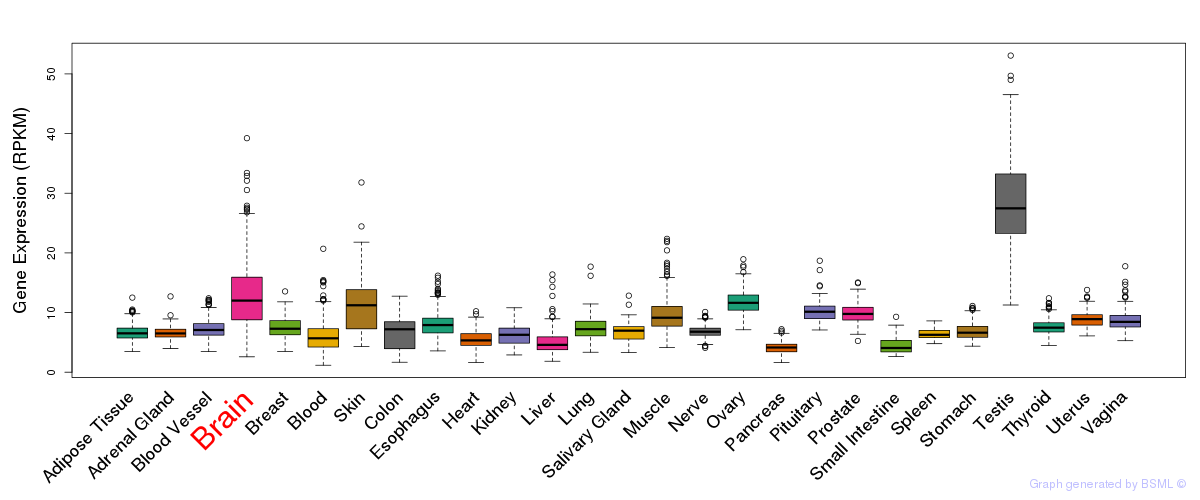
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| BRMS1L | 0.84 | 0.86 |
| FAM102B | 0.83 | 0.83 |
| OXCT1 | 0.81 | 0.81 |
| VAPB | 0.81 | 0.83 |
| EPS15 | 0.81 | 0.81 |
| USP12 | 0.80 | 0.80 |
| MCFD2 | 0.80 | 0.80 |
| SGTB | 0.80 | 0.80 |
| CDC40 | 0.80 | 0.78 |
| TRAPPC6B | 0.80 | 0.78 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| EFEMP2 | -0.39 | -0.46 |
| NUDT8 | -0.38 | -0.46 |
| NDUFA4L2 | -0.38 | -0.41 |
| EIF4EBP3 | -0.38 | -0.42 |
| AF347015.18 | -0.37 | -0.30 |
| GATSL3 | -0.37 | -0.43 |
| TMEM88 | -0.36 | -0.43 |
| MICALL2 | -0.35 | -0.37 |
| AP003068.3 | -0.35 | -0.34 |
| SLC38A5 | -0.35 | -0.42 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0050321 | tau-protein kinase activity | IDA | Brain (GO term level: 7) | 14594945 |
| GO:0050321 | tau-protein kinase activity | ISS | Brain (GO term level: 7) | - |
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 14594945 |14676191 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0005524 | ATP binding | NAS | 15009667 | |
| GO:0004674 | protein serine/threonine kinase activity | IDA | 15009667 | |
| GO:0004674 | protein serine/threonine kinase activity | IEA | - | |
| GO:0004674 | protein serine/threonine kinase activity | ISS | - | |
| GO:0004674 | protein serine/threonine kinase activity | NAS | 11326310 | |
| GO:0004713 | protein tyrosine kinase activity | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0008017 | microtubule binding | IDA | 14594945 | |
| GO:0008017 | microtubule binding | ISS | - | |
| GO:0043130 | ubiquitin binding | NAS | 14594945 | |
| GO:0043015 | gamma-tubulin binding | IDA | 14594945 | |
| GO:0043015 | gamma-tubulin binding | ISS | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007399 | nervous system development | IDA | neurite (GO term level: 5) | 14594945 |
| GO:0007399 | nervous system development | ISS | neurite (GO term level: 5) | - |
| GO:0000082 | G1/S transition of mitotic cell cycle | NAS | 12735302 | |
| GO:0000086 | G2/M transition of mitotic cell cycle | NAS | 11326310 | |
| GO:0001578 | microtubule bundle formation | IEP | 14594945 | |
| GO:0001578 | microtubule bundle formation | ISS | - | |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| GO:0006468 | protein amino acid phosphorylation | NAS | 11326310 | |
| GO:0006468 | protein amino acid phosphorylation | TAS | 14594945 | |
| GO:0016055 | Wnt receptor signaling pathway | TAS | 11326310 | |
| GO:0008284 | positive regulation of cell proliferation | IDA | 12735302 | |
| GO:0043068 | positive regulation of programmed cell death | NAS | 15009667 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0043005 | neuron projection | IDA | neuron, axon, neurite, dendrite (GO term level: 5) | 14594945 |
| GO:0043005 | neuron projection | ISS | neuron, axon, neurite, dendrite (GO term level: 5) | - |
| GO:0000930 | gamma-tubulin complex | IDA | 14594945 | |
| GO:0005813 | centrosome | IDA | 14594945 | |
| GO:0005813 | centrosome | ISS | - | |
| GO:0005815 | microtubule organizing center | IDA | 14594945 | |
| GO:0005874 | microtubule | IDA | 14594945 | |
| GO:0005737 | cytoplasm | IDA | 11326310 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTA1 | ACTA | ASMA | CFTD | CFTD1 | CFTDM | MPFD | NEM1 | NEM2 | NEM3 | actin, alpha 1, skeletal muscle | Affinity Capture-MS | BioGRID | 14594945 |
| HSPA4 | APG-2 | HS24/P52 | MGC131852 | RY | hsp70 | hsp70RY | heat shock 70kDa protein 4 | Affinity Capture-MS | BioGRID | 14594945 |
| MAP2 | DKFZp686I2148 | MAP2A | MAP2B | MAP2C | microtubule-associated protein 2 | - | HPRD,BioGRID | 14594945 |
| MAP4 | DKFZp779A1753 | MGC8617 | microtubule-associated protein 4 | - | HPRD,BioGRID | 14594945 |
| MAPT | DDPAC | FLJ31424 | FTDP-17 | MAPTL | MGC138549 | MSTD | MTBT1 | MTBT2 | PPND | TAU | microtubule-associated protein tau | - | HPRD,BioGRID | 14594945 |
| MYH10 | MGC134913 | MGC134914 | NMMHCB | myosin, heavy chain 10, non-muscle | Affinity Capture-MS | BioGRID | 14594945 |
| MYH9 | DFNA17 | EPSTS | FTNS | MGC104539 | MHA | NMHC-II-A | NMMHCA | myosin, heavy chain 9, non-muscle | Affinity Capture-MS | BioGRID | 14594945 |
| STK11 | LKB1 | PJS | serine/threonine kinase 11 | LKB1 phosphorylates MARK4. | BIND | 14676191 |
| TUBA1A | B-ALPHA-1 | FLJ25113 | LIS3 | TUBA3 | tubulin, alpha 1a | Affinity Capture-MS | BioGRID | 14594945 |
| TUBB | M40 | MGC117247 | MGC16435 | OK/SW-cl.56 | TUBB1 | TUBB5 | tubulin, beta | Affinity Capture-MS | BioGRID | 14594945 |
| TUBB2A | TUBB | TUBB2 | dJ40E16.7 | tubulin, beta 2A | - | HPRD | 14594945 |
| TUBG1 | TUBG | TUBGCP1 | tubulin, gamma 1 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 14594945 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID LKB1 PATHWAY | 47 | 37 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS UP | 473 | 314 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL DN | 275 | 168 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC UP | 185 | 126 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 8HR UP | 164 | 122 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| ROVERSI GLIOMA COPY NUMBER UP | 100 | 75 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM5 | 94 | 59 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS UP | 292 | 168 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL OPTIMAL DEBULKING | 246 | 152 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 36HR UP | 221 | 150 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 UP | 344 | 215 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-17-5p/20/93.mr/106/519.d | 759 | 765 | m8 | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-214 | 95 | 101 | m8 | hsa-miR-214brain | ACAGCAGGCACAGACAGGCAG |
| miR-24 | 269 | 276 | 1A,m8 | hsa-miR-24SZ | UGGCUCAGUUCAGCAGGAACAG |
| miR-493-5p | 765 | 771 | 1A | hsa-miR-493-5p | UUGUACAUGGUAGGCUUUCAUU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.