Gene Page: PTPRN2
Summary ?
| GeneID | 5799 |
| Symbol | PTPRN2 |
| Synonyms | IA-2beta|IAR|ICAAR|PTPRP|R-PTP-N2 |
| Description | protein tyrosine phosphatase, receptor type N2 |
| Reference | MIM:601698|HGNC:HGNC:9677|Ensembl:ENSG00000155093|HPRD:03413|Vega:OTTHUMG00000152646 |
| Gene type | protein-coding |
| Map location | 7q36 |
| Sherlock p-value | 0.051 |
| Fetal beta | -0.914 |
| DMG | 2 (# studies) |
| eGene | Meta |
| Support | TYROSINE KINASE SIGNALING CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 12 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 12 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg23220435 | 7 | 158107284 | PTPRN2 | 3.18E-6 | 0.415 | 0.009 | DMG:Wockner_2014 |
| cg08401938 | 7 | 158155932 | PTPRN2 | 8.85E-6 | 0.324 | 0.013 | DMG:Wockner_2014 |
| cg26389638 | 7 | 158264915 | PTPRN2 | 2.076E-4 | 0.374 | 0.035 | DMG:Wockner_2014 |
| cg09066883 | 7 | 157932951 | PTPRN2 | 2.289E-4 | 0.373 | 0.036 | DMG:Wockner_2014 |
| cg07213780 | 7 | 158342600 | PTPRN2 | 2.382E-4 | 0.46 | 0.037 | DMG:Wockner_2014 |
| cg09450352 | 7 | 157495481 | PTPRN2 | 2.612E-4 | 0.526 | 0.038 | DMG:Wockner_2014 |
| cg22881266 | 7 | 157543942 | PTPRN2 | 3.257E-4 | 0.326 | 0.04 | DMG:Wockner_2014 |
| cg26173773 | 7 | 158075624 | PTPRN2 | 3.337E-4 | 0.425 | 0.041 | DMG:Wockner_2014 |
| cg27448110 | 7 | 157484647 | PTPRN2 | 3.63E-4 | -0.41 | 0.042 | DMG:Wockner_2014 |
| cg18951543 | 7 | 157633867 | PTPRN2 | 3.95E-4 | -0.478 | 0.043 | DMG:Wockner_2014 |
| cg13632655 | 7 | 158266097 | PTPRN2 | 4.51E-4 | 0.406 | 0.045 | DMG:Wockner_2014 |
| cg25607226 | 7 | 157369960 | PTPRN2 | 7.6E-8 | 0.019 | 1.8E-5 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
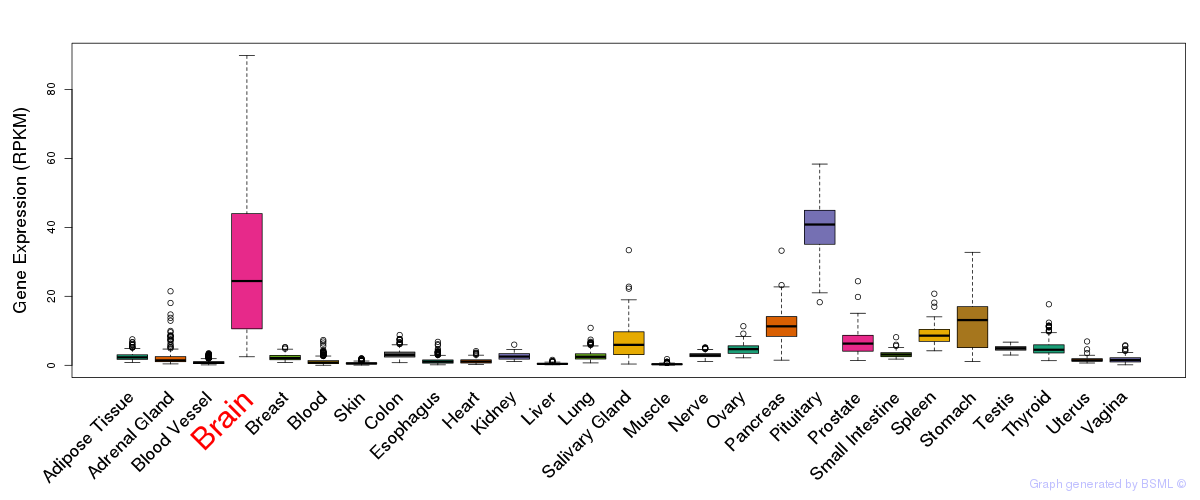
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NT5DC2 | 0.93 | 0.88 |
| PPP4C | 0.92 | 0.93 |
| SLC25A1 | 0.92 | 0.87 |
| SPNS1 | 0.91 | 0.90 |
| TSEN34 | 0.91 | 0.92 |
| WDR34 | 0.91 | 0.92 |
| DHRS13 | 0.91 | 0.91 |
| GPX7 | 0.90 | 0.85 |
| GNB2 | 0.90 | 0.92 |
| TARBP2 | 0.90 | 0.90 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.66 | -0.71 |
| AF347015.27 | -0.66 | -0.79 |
| HLA-F | -0.64 | -0.67 |
| AF347015.33 | -0.64 | -0.78 |
| AF347015.31 | -0.63 | -0.74 |
| MT-CO2 | -0.63 | -0.75 |
| AF347015.8 | -0.62 | -0.78 |
| TINAGL1 | -0.61 | -0.65 |
| MT-CYB | -0.60 | -0.76 |
| CA4 | -0.60 | -0.67 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG TYPE I DIABETES MELLITUS | 44 | 38 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA DN | 349 | 157 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER ADVANCED VS EARLY DN | 138 | 70 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 AND HDAC2 TARGETS UP | 238 | 144 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| CHOW RASSF1 TARGETS DN | 29 | 19 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 1 UP | 121 | 71 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 DN | 229 | 142 | All SZGR 2.0 genes in this pathway |
| VETTER TARGETS OF PRKCA AND ETS1 DN | 16 | 10 | All SZGR 2.0 genes in this pathway |
| ROVERSI GLIOMA COPY NUMBER UP | 100 | 75 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER DN | 203 | 134 | All SZGR 2.0 genes in this pathway |
| KIM MYC AMPLIFICATION TARGETS DN | 97 | 51 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN | 637 | 377 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| SASAKI ADULT T CELL LEUKEMIA | 176 | 122 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE UP | 244 | 165 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC UP | 202 | 115 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY 4NQO OR UV | 63 | 44 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE UP | 226 | 164 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| EHLERS ANEUPLOIDY UP | 41 | 29 | All SZGR 2.0 genes in this pathway |
| TAVAZOIE METASTASIS | 108 | 68 | All SZGR 2.0 genes in this pathway |
| MONTERO THYROID CANCER POOR SURVIVAL DN | 11 | 6 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 AND H3K27ME3 | 349 | 234 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS DN | 306 | 191 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K4ME3 AND H3K27ME3 | 210 | 148 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| ALFANO MYC TARGETS | 239 | 156 | All SZGR 2.0 genes in this pathway |
| RATTENBACHER BOUND BY CELF1 | 467 | 251 | All SZGR 2.0 genes in this pathway |