Gene Page: BBS4
Summary ?
| GeneID | 585 |
| Symbol | BBS4 |
| Synonyms | - |
| Description | Bardet-Biedl syndrome 4 |
| Reference | MIM:600374|HGNC:HGNC:969|Ensembl:ENSG00000140463|HPRD:08982|Vega:OTTHUMG00000133510 |
| Gene type | protein-coding |
| Map location | 15q22.3-q23 |
| Pascal p-value | 0.861 |
| Sherlock p-value | 0.281 |
| Fetal beta | -0.393 |
| eGene | Cerebellar Hemisphere Cerebellum Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs12594938 | chr15 | 73071281 | BBS4 | 585 | 0.06 | cis | ||
| rs10770856 | chr12 | 8705405 | BBS4 | 585 | 0.12 | trans |
Section II. Transcriptome annotation
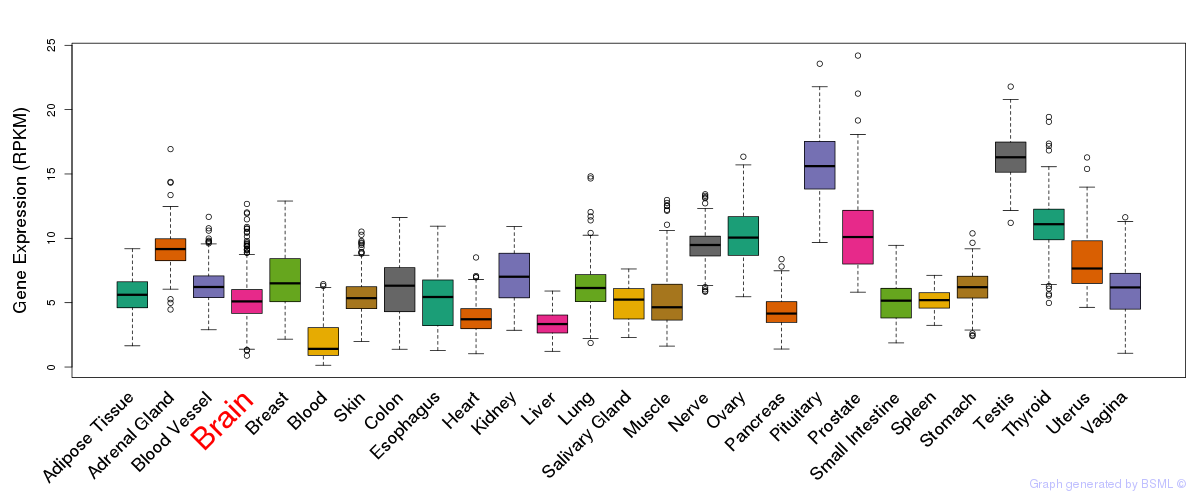
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003777 | microtubule motor activity | IMP | 15107855 | |
| GO:0005515 | protein binding | IPI | 15107855 | |
| GO:0034452 | dynactin binding | IDA | 15107855 | |
| GO:0043014 | alpha-tubulin binding | IDA | 17574030 | |
| GO:0048487 | beta-tubulin binding | IDA | 17574030 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0035058 | sensory cilium biogenesis | IEA | neuron (GO term level: 10) | - |
| GO:0016358 | dendrite development | IEA | neurite, dendrite (GO term level: 11) | - |
| GO:0000226 | microtubule cytoskeleton organization | IEA | - | |
| GO:0001895 | retina homeostasis | IEA | - | |
| GO:0001843 | neural tube closure | IEA | - | |
| GO:0007286 | spermatid development | IEA | - | |
| GO:0007608 | sensory perception of smell | IEA | - | |
| GO:0033365 | protein localization in organelle | ISS | - | |
| GO:0033205 | cytokinesis during cell cycle | IMP | 15107855 | |
| GO:0034454 | microtubule anchoring at centrosome | IMP | 15107855 | |
| GO:0030534 | adult behavior | IEA | - | |
| GO:0050896 | response to stimulus | IEA | - | |
| GO:0050893 | sensory processing | TAS | 14520415 | |
| GO:0019216 | regulation of lipid metabolic process | IEA | - | |
| GO:0051297 | centrosome organization | IMP | 15107855 | |
| GO:0032465 | regulation of cytokinesis | IMP | 15107855 | |
| GO:0045724 | positive regulation of flagellum biogenesis | IEA | - | |
| GO:0046548 | retinal rod cell development | IEA | - | |
| GO:0045494 | photoreceptor cell maintenance | IEA | - | |
| GO:0051457 | maintenance of protein location in nucleus | IGI | 15107855 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000242 | pericentriolar material | IDA | 15107855 | |
| GO:0005814 | centriole | IDA | 15107855 | |
| GO:0005856 | cytoskeleton | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005932 | basal body | IDA | 15107855 |18299575 | |
| GO:0034464 | BBSome | IDA | 17574030 | |
| GO:0034451 | centriolar satellite | IDA | 15107855 | |
| GO:0031514 | motile secondary cilium | IDA | 18299575 | |
| GO:0031513 | nonmotile primary cilium | IDA | 17574030 | |
| GO:0060170 | cilium membrane | IDA | 17574030 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| CHEMNITZ RESPONSE TO PROSTAGLANDIN E2 DN | 391 | 222 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS UP | 214 | 155 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 12HR DN | 209 | 122 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| YANG BREAST CANCER ESR1 UP | 36 | 22 | All SZGR 2.0 genes in this pathway |
| NOUZOVA TRETINOIN AND H4 ACETYLATION | 143 | 85 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS EARLY PROGENITOR | 532 | 309 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR DN | 101 | 70 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| YEGNASUBRAMANIAN PROSTATE CANCER | 128 | 60 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE UP | 578 | 341 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| MATZUK SPERMATOZOA | 114 | 77 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 1HR DN | 222 | 147 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 UP | 344 | 215 | All SZGR 2.0 genes in this pathway |
| KRIEG KDM3A TARGETS NOT HYPOXIA | 208 | 107 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |
| SMIRNOV RESPONSE TO IR 6HR UP | 166 | 97 | All SZGR 2.0 genes in this pathway |