Gene Page: KDM5A
Summary ?
| GeneID | 5927 |
| Symbol | KDM5A |
| Synonyms | RBBP-2|RBBP2|RBP2 |
| Description | lysine demethylase 5A |
| Reference | MIM:180202|HGNC:HGNC:9886|Ensembl:ENSG00000073614|HPRD:08915|Vega:OTTHUMG00000168055 |
| Gene type | protein-coding |
| Map location | 12p11 |
| Pascal p-value | 0.586 |
| Sherlock p-value | 0.874 |
| Fetal beta | 0.38 |
| Support | Ascano FMRP targets Chromatin Remodeling Genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
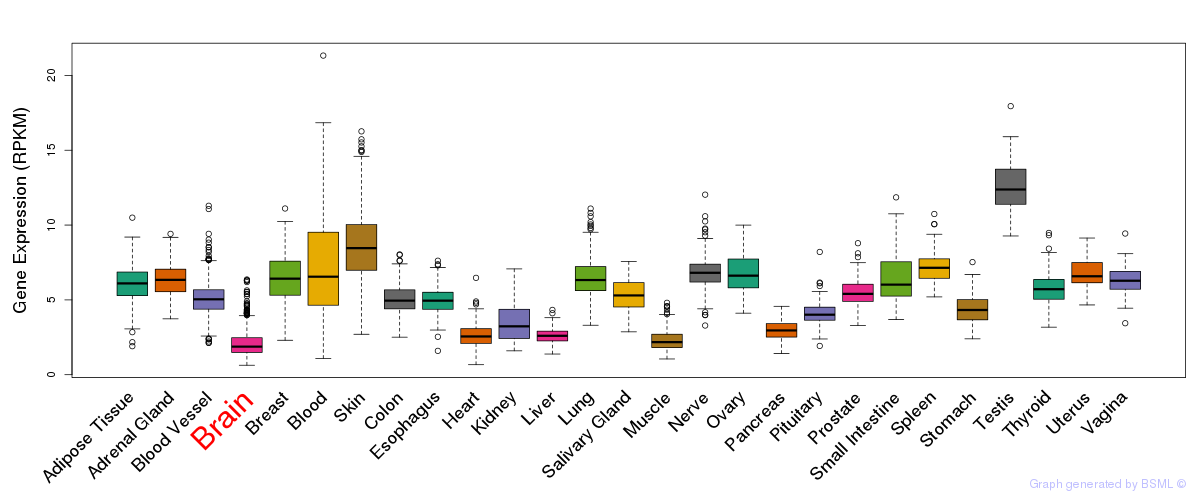
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HEXDC | 0.84 | 0.84 |
| PCBP4 | 0.81 | 0.84 |
| ACD | 0.81 | 0.77 |
| UCKL1 | 0.81 | 0.81 |
| FBXL6 | 0.81 | 0.79 |
| CLN3 | 0.80 | 0.81 |
| PLSCR3 | 0.80 | 0.82 |
| ZBTB48 | 0.79 | 0.79 |
| RFNG | 0.79 | 0.80 |
| QTRT1 | 0.79 | 0.82 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.27 | -0.56 | -0.59 |
| AF347015.31 | -0.54 | -0.58 |
| MT-CO2 | -0.52 | -0.57 |
| MT-CYB | -0.52 | -0.57 |
| AF347015.8 | -0.52 | -0.58 |
| AF347015.15 | -0.51 | -0.58 |
| MT-ATP8 | -0.49 | -0.64 |
| AF347015.33 | -0.49 | -0.53 |
| ABCG2 | -0.49 | -0.52 |
| AF347015.21 | -0.48 | -0.62 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BGLAP | BGP | OC | PMF1 | bone gamma-carboxyglutamate (gla) protein | RBP2 interacts with OC promoter. | BIND | 15949438 |
| BRD2 | D6S113E | DKFZp686N0336 | FLJ31942 | FSH | FSRG1 | KIAA9001 | NAT | RING3 | RNF3 | bromodomain containing 2 | RBP2 interacts with BRD2 promoter. | BIND | 15949438 |
| BRD8 | SMAP | SMAP2 | p120 | bromodomain containing 8 | RBP2 interacts with BRD8 promoter. | BIND | 15949438 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 11358960 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | Rbp2 interacts with ER. | BIND | 11358960 |
| LMO2 | RBTN2 | RBTNL1 | RHOM2 | TTG2 | LIM domain only 2 (rhombotin-like 1) | - | HPRD,BioGRID | 9129143 |
| NR3C1 | GCCR | GCR | GR | GRL | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) | Reconstituted Complex | BioGRID | 11358960 |
| NR3C1 | GCCR | GCR | GR | GRL | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) | Rbp2 interacts with GR. This interaction was modeled on a demonstrated interaction between human Rbp2 and rat GR. | BIND | 11358960 |
| RARA | NR1B1 | RAR | retinoic acid receptor, alpha | Reconstituted Complex | BioGRID | 11358960 |
| RARA | NR1B1 | RAR | retinoic acid receptor, alpha | Rbp2 interacts with RAR. | BIND | 11358960 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | - | HPRD,BioGRID | 7935440 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | Rbp2 interacts with Rb. This interaction was modeled on a demonstrated interaction between human Rbp2 and Rb from an unspecified species. | BIND | 11358960 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | pRB interacts with RBP2. | BIND | 15949438 |
| RBL1 | CP107 | MGC40006 | PRB1 | p107 | retinoblastoma-like 1 (p107) | - | HPRD,BioGRID | 7935440 |
| TBP | GTF2D | GTF2D1 | MGC117320 | MGC126054 | MGC126055 | SCA17 | TFIID | TATA box binding protein | - | HPRD,BioGRID | 7935440 |
| VDR | NR1I1 | vitamin D (1,25- dihydroxyvitamin D3) receptor | - | HPRD,BioGRID | 11358960 |
| VDR | NR1I1 | vitamin D (1,25- dihydroxyvitamin D3) receptor | Rbp2 interacts with VDR. | BIND | 11358960 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 5 6WK DN | 137 | 97 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 6 7WK UP | 197 | 135 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION HBZ | 41 | 27 | All SZGR 2.0 genes in this pathway |
| JAERVINEN AMPLIFIED IN LARYNGEAL CANCER | 40 | 24 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST DN | 309 | 206 | All SZGR 2.0 genes in this pathway |
| HOFMANN CELL LYMPHOMA UP | 50 | 35 | All SZGR 2.0 genes in this pathway |
| NATSUME RESPONSE TO INTERFERON BETA UP | 71 | 49 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| JAZAERI BREAST CANCER BRCA1 VS BRCA2 UP | 49 | 28 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK DN | 318 | 220 | All SZGR 2.0 genes in this pathway |
| TRACEY RESISTANCE TO IFNA2 DN | 32 | 23 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| CHO NR4A1 TARGETS | 33 | 22 | All SZGR 2.0 genes in this pathway |
| LEE DIFFERENTIATING T LYMPHOCYTE | 200 | 115 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS UP | 221 | 135 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 SIGNALING VIA NFIC 1HR DN | 106 | 77 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 SIGNALING VIA NFIC 10HR UP | 54 | 38 | All SZGR 2.0 genes in this pathway |