Gene Page: ELAC2
Summary ?
| GeneID | 60528 |
| Symbol | ELAC2 |
| Synonyms | COXPD17|ELC2|HPC2 |
| Description | elaC ribonuclease Z 2 |
| Reference | MIM:605367|HGNC:HGNC:14198|HPRD:05641| |
| Gene type | protein-coding |
| Map location | 17p11.2 |
| Pascal p-value | 2.13E-4 |
| Sherlock p-value | 0.412 |
| Fetal beta | -0.326 |
| DMG | 1 (# studies) |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Cerebellar Hemisphere Cerebellum Cortex Frontal Cortex BA9 Hippocampus Hypothalamus Nucleus accumbens basal ganglia Putamen basal ganglia |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg08011170 | 17 | 12921393 | ELAC2 | 2.07E-9 | -0.023 | 1.66E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2523 | 17 | 12895815 | ELAC2 | ENSG00000006744.14 | 4.545E-10 | 0 | 25689 | gtex_brain_ba24 |
| rs1044569 | 17 | 12895929 | ELAC2 | ENSG00000006744.14 | 4.545E-10 | 0 | 25575 | gtex_brain_ba24 |
| rs1044564 | 17 | 12895984 | ELAC2 | ENSG00000006744.14 | 2.491E-10 | 0 | 25520 | gtex_brain_ba24 |
| rs9908102 | 17 | 12896553 | ELAC2 | ENSG00000006744.14 | 4.529E-10 | 0 | 24951 | gtex_brain_ba24 |
| rs9915058 | 17 | 12897293 | ELAC2 | ENSG00000006744.14 | 2.98E-7 | 0 | 24211 | gtex_brain_ba24 |
| rs62058145 | 17 | 12913490 | ELAC2 | ENSG00000006744.14 | 9.182E-9 | 0 | 8014 | gtex_brain_ba24 |
| rs11658321 | 17 | 12914484 | ELAC2 | ENSG00000006744.14 | 3.632E-8 | 0 | 7020 | gtex_brain_ba24 |
| rs2523 | 17 | 12895815 | ELAC2 | ENSG00000006744.14 | 8.95E-8 | 0.01 | 25689 | gtex_brain_putamen_basal |
| rs1044569 | 17 | 12895929 | ELAC2 | ENSG00000006744.14 | 1.036E-7 | 0.01 | 25575 | gtex_brain_putamen_basal |
| rs9908102 | 17 | 12896553 | ELAC2 | ENSG00000006744.14 | 1.11E-6 | 0.01 | 24951 | gtex_brain_putamen_basal |
| rs11658321 | 17 | 12914484 | ELAC2 | ENSG00000006744.14 | 7.601E-7 | 0.01 | 7020 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
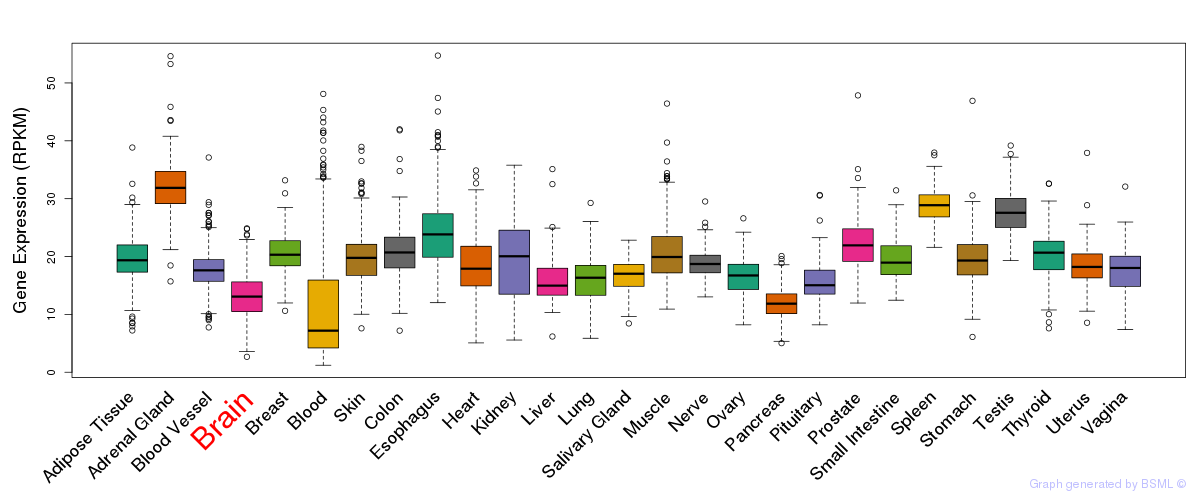
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS UP | 293 | 179 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| FERREIRA EWINGS SARCOMA UNSTABLE VS STABLE DN | 98 | 59 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR UP | 180 | 125 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE DN | 261 | 183 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE UP | 393 | 244 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS DN | 292 | 189 | All SZGR 2.0 genes in this pathway |
| MUELLER PLURINET | 299 | 189 | All SZGR 2.0 genes in this pathway |
| ICHIBA GRAFT VERSUS HOST DISEASE 35D DN | 49 | 34 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |