Gene Page: CCL3
Summary ?
| GeneID | 6348 |
| Symbol | CCL3 |
| Synonyms | G0S19-1|LD78ALPHA|MIP-1-alpha|MIP1A|SCYA3 |
| Description | C-C motif chemokine ligand 3 |
| Reference | MIM:182283|HGNC:HGNC:10627|Ensembl:ENSG00000277632|HPRD:01656|Vega:OTTHUMG00000188413 |
| Gene type | protein-coding |
| Map location | 17q12 |
| Pascal p-value | 0.539 |
| Fetal beta | -0.223 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10491115 | chr17 | 33969406 | CCL3 | 6348 | 0.1 | cis | ||
| rs3760325 | chr17 | 34037419 | CCL3 | 6348 | 0.08 | cis | ||
| rs983619 | chr1 | 39047418 | CCL3 | 6348 | 0.12 | trans | ||
| rs7545984 | chr1 | 45003892 | CCL3 | 6348 | 0.15 | trans | ||
| rs7554123 | chr1 | 45004203 | CCL3 | 6348 | 0.15 | trans | ||
| rs12059674 | chr1 | 57630277 | CCL3 | 6348 | 0.11 | trans | ||
| rs17474410 | chr1 | 68267242 | CCL3 | 6348 | 0.18 | trans | ||
| rs7511937 | 0 | CCL3 | 6348 | 0.06 | trans | |||
| rs17689089 | chr1 | 77740105 | CCL3 | 6348 | 0 | trans | ||
| rs17100231 | chr1 | 77761339 | CCL3 | 6348 | 0.06 | trans | ||
| rs17100274 | chr1 | 77774412 | CCL3 | 6348 | 1.08E-4 | trans | ||
| rs12034005 | chr1 | 79793034 | CCL3 | 6348 | 0.14 | trans | ||
| rs17098656 | chr1 | 80098386 | CCL3 | 6348 | 0 | trans | ||
| rs6664544 | chr1 | 92524192 | CCL3 | 6348 | 0.1 | trans | ||
| rs12403481 | chr1 | 109503655 | CCL3 | 6348 | 0 | trans | ||
| rs337294 | 0 | CCL3 | 6348 | 0.01 | trans | |||
| rs17029293 | chr1 | 112567844 | CCL3 | 6348 | 8.653E-6 | trans | ||
| rs12568798 | chr1 | 164355032 | CCL3 | 6348 | 0.07 | trans | ||
| rs6701323 | chr1 | 178588280 | CCL3 | 6348 | 0.19 | trans | ||
| rs16843112 | chr1 | 198420657 | CCL3 | 6348 | 0.15 | trans | ||
| rs697455 | chr1 | 202512075 | CCL3 | 6348 | 0.08 | trans | ||
| rs1796829 | chr1 | 239661818 | CCL3 | 6348 | 0.02 | trans | ||
| rs12034521 | chr1 | 239717679 | CCL3 | 6348 | 0 | trans | ||
| snp_a-1990354 | 0 | CCL3 | 6348 | 3.783E-5 | trans | |||
| rs6432434 | chr2 | 14160930 | CCL3 | 6348 | 0.13 | trans | ||
| rs12329305 | chr2 | 19552912 | CCL3 | 6348 | 0.07 | trans | ||
| rs12611832 | chr2 | 31196248 | CCL3 | 6348 | 5.841E-5 | trans | ||
| rs12614732 | chr2 | 31199913 | CCL3 | 6348 | 5.841E-5 | trans | ||
| rs7560081 | chr2 | 62793238 | CCL3 | 6348 | 0.16 | trans | ||
| rs17029079 | chr2 | 64736071 | CCL3 | 6348 | 0.07 | trans | ||
| rs17038084 | chr2 | 85192280 | CCL3 | 6348 | 1.271E-5 | trans | ||
| rs1840209 | chr2 | 125625401 | CCL3 | 6348 | 0.08 | trans | ||
| rs6707117 | chr2 | 136855504 | CCL3 | 6348 | 1.201E-7 | trans | ||
| rs10189091 | chr2 | 143471999 | CCL3 | 6348 | 0.01 | trans | ||
| snp_a-2198363 | 0 | CCL3 | 6348 | 0.09 | trans | |||
| rs707097 | chr2 | 155318814 | CCL3 | 6348 | 0.02 | trans | ||
| rs16838911 | chr2 | 156087569 | CCL3 | 6348 | 8.507E-4 | trans | ||
| rs7608942 | chr2 | 182820100 | CCL3 | 6348 | 0.05 | trans | ||
| rs16829292 | chr2 | 233447622 | CCL3 | 6348 | 0.07 | trans | ||
| rs16829401 | chr2 | 233538391 | CCL3 | 6348 | 0.07 | trans | ||
| rs17040622 | chr3 | 4440673 | CCL3 | 6348 | 0.19 | trans | ||
| rs17684934 | chr3 | 4443547 | CCL3 | 6348 | 0.01 | trans | ||
| rs17040633 | chr3 | 4446836 | CCL3 | 6348 | 0.01 | trans | ||
| rs17756483 | chr3 | 4451309 | CCL3 | 6348 | 0.01 | trans | ||
| rs17685441 | chr3 | 4465165 | CCL3 | 6348 | 0.01 | trans | ||
| rs3796213 | chr3 | 69157380 | CCL3 | 6348 | 0.14 | trans | ||
| rs2875492 | chr3 | 86006209 | CCL3 | 6348 | 0.08 | trans | ||
| rs16843128 | chr3 | 100713413 | CCL3 | 6348 | 0.08 | trans | ||
| rs1381835 | chr3 | 110600595 | CCL3 | 6348 | 9.419E-4 | trans | ||
| rs9288980 | chr3 | 112964991 | CCL3 | 6348 | 0.01 | trans | ||
| rs3995908 | chr3 | 112998529 | CCL3 | 6348 | 0.01 | trans | ||
| rs7652801 | chr3 | 113008605 | CCL3 | 6348 | 0.01 | trans | ||
| rs7625829 | chr3 | 113103743 | CCL3 | 6348 | 0.01 | trans | ||
| rs4419353 | chr3 | 113254936 | CCL3 | 6348 | 0.15 | trans | ||
| rs17636599 | chr3 | 143028186 | CCL3 | 6348 | 0.01 | trans | ||
| rs16839796 | chr4 | 7188348 | CCL3 | 6348 | 4.118E-4 | trans | ||
| rs16885724 | chr4 | 31717283 | CCL3 | 6348 | 0.2 | trans | ||
| rs11096966 | chr4 | 38961258 | CCL3 | 6348 | 0.12 | trans | ||
| rs4469113 | chr4 | 41461062 | CCL3 | 6348 | 0.01 | trans | ||
| rs6836259 | chr4 | 57173074 | CCL3 | 6348 | 0.08 | trans | ||
| rs2643038 | chr4 | 62313218 | CCL3 | 6348 | 0.19 | trans | ||
| rs6817626 | chr4 | 78005836 | CCL3 | 6348 | 0.17 | trans | ||
| rs1604153 | chr4 | 78472744 | CCL3 | 6348 | 0.16 | trans | ||
| rs1564462 | chr4 | 79910988 | CCL3 | 6348 | 0.03 | trans | ||
| rs17005000 | chr4 | 81826551 | CCL3 | 6348 | 0.01 | trans | ||
| rs10011751 | chr4 | 93984994 | CCL3 | 6348 | 0.03 | trans | ||
| rs6881821 | chr5 | 8578465 | CCL3 | 6348 | 0.01 | trans | ||
| rs1697073 | chr5 | 30907267 | CCL3 | 6348 | 0.09 | trans | ||
| rs2115110 | chr5 | 38193428 | CCL3 | 6348 | 0.01 | trans | ||
| rs16873956 | chr5 | 44312488 | CCL3 | 6348 | 0.1 | trans | ||
| rs10040715 | chr5 | 57594207 | CCL3 | 6348 | 8.691E-8 | trans | ||
| rs16894149 | chr5 | 61013724 | CCL3 | 6348 | 0.01 | trans | ||
| snp_a-4217300 | 0 | CCL3 | 6348 | 5.64E-8 | trans | |||
| rs163953 | chr5 | 92289536 | CCL3 | 6348 | 0.12 | trans | ||
| rs17134275 | chr5 | 100634998 | CCL3 | 6348 | 0.15 | trans | ||
| rs26604 | chr5 | 115792367 | CCL3 | 6348 | 0.07 | trans | ||
| rs1350578 | chr5 | 116310282 | CCL3 | 6348 | 0.15 | trans | ||
| rs2141482 | chr5 | 136760477 | CCL3 | 6348 | 0.01 | trans | ||
| rs2189803 | chr5 | 137684661 | CCL3 | 6348 | 7.546E-5 | trans | ||
| rs17063216 | chr5 | 163777647 | CCL3 | 6348 | 1.644E-4 | trans | ||
| rs17063224 | chr5 | 163780687 | CCL3 | 6348 | 1.644E-4 | trans | ||
| rs259913 | chr5 | 169370782 | CCL3 | 6348 | 0.01 | trans | ||
| rs2116811 | chr5 | 171820051 | CCL3 | 6348 | 0.06 | trans | ||
| rs258875 | chr5 | 173046554 | CCL3 | 6348 | 0.01 | trans | ||
| rs16883859 | chr6 | 20474617 | CCL3 | 6348 | 0.05 | trans | ||
| rs16883862 | chr6 | 20474766 | CCL3 | 6348 | 0.05 | trans | ||
| rs10484635 | chr6 | 20538664 | CCL3 | 6348 | 0.19 | trans | ||
| rs2057537 | chr6 | 35090278 | CCL3 | 6348 | 0.17 | trans | ||
| rs195429 | chr6 | 37354434 | CCL3 | 6348 | 0.05 | trans | ||
| rs12528414 | chr6 | 38404800 | CCL3 | 6348 | 0.08 | trans | ||
| rs7771164 | chr6 | 40442772 | CCL3 | 6348 | 0.03 | trans | ||
| rs4289676 | chr6 | 123999302 | CCL3 | 6348 | 0.15 | trans | ||
| rs17055235 | chr6 | 128355263 | CCL3 | 6348 | 0.02 | trans | ||
| rs12176346 | chr6 | 136732663 | CCL3 | 6348 | 0.02 | trans | ||
| rs9402819 | chr6 | 136848169 | CCL3 | 6348 | 0.09 | trans | ||
| rs9389400 | chr6 | 136854803 | CCL3 | 6348 | 0.09 | trans | ||
| rs2237264 | chr6 | 136919977 | CCL3 | 6348 | 0.02 | trans | ||
| rs17080121 | chr6 | 151009189 | CCL3 | 6348 | 0.06 | trans | ||
| rs12529269 | chr6 | 153543829 | CCL3 | 6348 | 0.14 | trans | ||
| rs10872699 | chr6 | 153544252 | CCL3 | 6348 | 0.01 | trans | ||
| rs1321606 | chr6 | 153545278 | CCL3 | 6348 | 0.08 | trans | ||
| rs10945911 | chr6 | 164047509 | CCL3 | 6348 | 0.01 | trans | ||
| rs17136841 | chr7 | 16963043 | CCL3 | 6348 | 0.15 | trans | ||
| rs12540348 | chr7 | 25802146 | CCL3 | 6348 | 0.04 | trans | ||
| rs4719825 | chr7 | 25813917 | CCL3 | 6348 | 0 | trans | ||
| rs6461881 | chr7 | 25837688 | CCL3 | 6348 | 0.05 | trans | ||
| rs13239699 | chr7 | 26377619 | CCL3 | 6348 | 0.08 | trans | ||
| rs12701006 | chr7 | 30147298 | CCL3 | 6348 | 1.785E-5 | trans | ||
| rs17621151 | chr7 | 53572911 | CCL3 | 6348 | 8.16E-4 | trans | ||
| rs10245449 | chr7 | 80935100 | CCL3 | 6348 | 0.18 | trans | ||
| rs17166937 | chr7 | 95179901 | CCL3 | 6348 | 0.11 | trans | ||
| rs2272618 | chr7 | 107871360 | CCL3 | 6348 | 0.11 | trans | ||
| rs12669568 | chr7 | 107885942 | CCL3 | 6348 | 0.11 | trans | ||
| rs7777322 | chr7 | 107886663 | CCL3 | 6348 | 0.11 | trans | ||
| rs6958821 | chr7 | 153253566 | CCL3 | 6348 | 1.208E-5 | trans | ||
| rs11779237 | chr8 | 1227664 | CCL3 | 6348 | 0.06 | trans | ||
| rs1010895 | chr8 | 14580230 | CCL3 | 6348 | 0.19 | trans | ||
| rs7001738 | chr8 | 26455799 | CCL3 | 6348 | 0.08 | trans | ||
| rs16877914 | chr8 | 31196384 | CCL3 | 6348 | 0.15 | trans | ||
| rs1457044 | chr8 | 59257567 | CCL3 | 6348 | 2.343E-4 | trans | ||
| rs1457032 | chr8 | 59290536 | CCL3 | 6348 | 0.01 | trans | ||
| rs2680903 | chr8 | 59512333 | CCL3 | 6348 | 0.13 | trans | ||
| rs2634533 | chr8 | 59535621 | CCL3 | 6348 | 0.18 | trans | ||
| rs7465259 | 0 | CCL3 | 6348 | 0 | trans | |||
| rs10104622 | chr8 | 63333773 | CCL3 | 6348 | 0 | trans | ||
| rs2218911 | chr8 | 63371015 | CCL3 | 6348 | 0 | trans | ||
| rs1551199 | chr8 | 63388117 | CCL3 | 6348 | 0 | trans | ||
| rs1593374 | chr8 | 64845850 | CCL3 | 6348 | 0 | trans | ||
| rs1733950 | chr8 | 90062516 | CCL3 | 6348 | 3.125E-9 | trans | ||
| rs1240028 | chr8 | 90093357 | CCL3 | 6348 | 0 | trans | ||
| rs1240087 | chr8 | 90113945 | CCL3 | 6348 | 0.01 | trans | ||
| rs318307 | chr8 | 90185459 | CCL3 | 6348 | 0 | trans | ||
| rs1483373 | chr8 | 90186442 | CCL3 | 6348 | 0.07 | trans | ||
| rs11776005 | chr8 | 90187416 | CCL3 | 6348 | 0.02 | trans | ||
| rs1159499 | chr8 | 90196948 | CCL3 | 6348 | 0.07 | trans | ||
| rs1355060 | chr8 | 118717710 | CCL3 | 6348 | 0.14 | trans | ||
| rs2280828 | chr8 | 118762435 | CCL3 | 6348 | 0.11 | trans | ||
| rs2514602 | chr8 | 119650549 | CCL3 | 6348 | 0.13 | trans | ||
| rs17683401 | chr8 | 119671834 | CCL3 | 6348 | 0.05 | trans | ||
| rs4976941 | chr8 | 142744439 | CCL3 | 6348 | 0.01 | trans | ||
| rs9298651 | chr9 | 10076275 | CCL3 | 6348 | 0.01 | trans | ||
| rs16931013 | chr9 | 10076859 | CCL3 | 6348 | 0.01 | trans | ||
| rs16931018 | chr9 | 10077048 | CCL3 | 6348 | 0.01 | trans | ||
| rs1467547 | chr9 | 17630726 | CCL3 | 6348 | 0.06 | trans | ||
| rs2210327 | chr9 | 18109234 | CCL3 | 6348 | 0.04 | trans | ||
| rs6476383 | chr9 | 32865568 | CCL3 | 6348 | 0.03 | trans | ||
| rs11143622 | chr9 | 71370050 | CCL3 | 6348 | 0.01 | trans | ||
| rs11143868 | chr9 | 71437411 | CCL3 | 6348 | 0.12 | trans | ||
| rs11143995 | chr9 | 71462852 | CCL3 | 6348 | 0.12 | trans | ||
| rs11144031 | chr9 | 71477107 | CCL3 | 6348 | 0.12 | trans | ||
| rs11791891 | chr9 | 87512411 | CCL3 | 6348 | 0.13 | trans | ||
| rs2165893 | chr9 | 87531423 | CCL3 | 6348 | 0.11 | trans | ||
| rs3780642 | chr9 | 87553144 | CCL3 | 6348 | 2.357E-4 | trans | ||
| rs10992752 | chr9 | 96240578 | CCL3 | 6348 | 0.11 | trans | ||
| rs2807706 | chr9 | 121744331 | CCL3 | 6348 | 0.02 | trans | ||
| rs2818297 | chr9 | 121757015 | CCL3 | 6348 | 0.1 | trans | ||
| rs10739527 | chr9 | 121772512 | CCL3 | 6348 | 0.17 | trans | ||
| rs11103132 | chr9 | 138575901 | CCL3 | 6348 | 0.12 | trans | ||
| rs11014370 | chr10 | 25352798 | CCL3 | 6348 | 0.06 | trans | ||
| rs17432714 | chr10 | 43526022 | CCL3 | 6348 | 0.1 | trans | ||
| rs2853836 | chr10 | 48431948 | CCL3 | 6348 | 0.19 | trans | ||
| rs3104871 | chr10 | 57878502 | CCL3 | 6348 | 0 | trans | ||
| rs901309 | chr10 | 72571899 | CCL3 | 6348 | 0.14 | trans | ||
| rs17271314 | chr10 | 86662514 | CCL3 | 6348 | 0.02 | trans | ||
| rs485305 | chr10 | 116048044 | CCL3 | 6348 | 6.179E-4 | trans | ||
| rs4750827 | chr10 | 132547183 | CCL3 | 6348 | 0.05 | trans | ||
| rs4750829 | chr10 | 132547260 | CCL3 | 6348 | 0.08 | trans | ||
| rs11021900 | chr11 | 11548236 | CCL3 | 6348 | 0.18 | trans | ||
| rs862785 | chr11 | 20175604 | CCL3 | 6348 | 0.03 | trans | ||
| rs16911034 | chr11 | 23302320 | CCL3 | 6348 | 0.09 | trans | ||
| rs11020043 | chr11 | 92470587 | CCL3 | 6348 | 0.1 | trans | ||
| rs1789300 | chr11 | 95463631 | CCL3 | 6348 | 0.01 | trans | ||
| rs1727152 | chr11 | 95467435 | CCL3 | 6348 | 0.04 | trans | ||
| rs1255149 | chr11 | 95484493 | CCL3 | 6348 | 0.01 | trans | ||
| rs3748251 | chr11 | 95519156 | CCL3 | 6348 | 0 | trans | ||
| rs16922543 | chr11 | 95526116 | CCL3 | 6348 | 0 | trans | ||
| rs16922622 | chr11 | 95567387 | CCL3 | 6348 | 0 | trans | ||
| rs3181323 | chr11 | 104878386 | CCL3 | 6348 | 0.02 | trans | ||
| rs11219092 | chr11 | 123130750 | CCL3 | 6348 | 0.01 | trans | ||
| rs374484 | chr12 | 27627159 | CCL3 | 6348 | 0.08 | trans | ||
| rs10783318 | chr12 | 39070575 | CCL3 | 6348 | 0.05 | trans | ||
| rs180424 | chr12 | 47171757 | CCL3 | 6348 | 0.13 | trans | ||
| rs17097455 | chr12 | 47398974 | CCL3 | 6348 | 0.12 | trans | ||
| rs17097566 | chr12 | 47460199 | CCL3 | 6348 | 0.12 | trans | ||
| rs8490 | chr12 | 51453904 | CCL3 | 6348 | 6.239E-5 | trans | ||
| rs1344796 | chr12 | 76397104 | CCL3 | 6348 | 0.08 | trans | ||
| rs10850201 | chr12 | 109903061 | CCL3 | 6348 | 0.15 | trans | ||
| rs11833075 | chr12 | 122114592 | CCL3 | 6348 | 0 | trans | ||
| rs1041050 | chr13 | 26585121 | CCL3 | 6348 | 0.02 | trans | ||
| rs2408301 | chr13 | 51808915 | CCL3 | 6348 | 2.113E-4 | trans | ||
| rs7982110 | chr13 | 59723428 | CCL3 | 6348 | 0.01 | trans | ||
| rs7330331 | chr13 | 67403122 | CCL3 | 6348 | 0.02 | trans | ||
| rs17088392 | chr13 | 72257235 | CCL3 | 6348 | 0 | trans | ||
| rs7327697 | chr13 | 83227456 | CCL3 | 6348 | 0.01 | trans | ||
| rs17667894 | chr13 | 92014308 | CCL3 | 6348 | 0.1 | trans | ||
| rs17257605 | chr14 | 25405534 | CCL3 | 6348 | 0.04 | trans | ||
| rs7156088 | chr14 | 27821865 | CCL3 | 6348 | 0.04 | trans | ||
| rs17106703 | chr14 | 37851979 | CCL3 | 6348 | 0.08 | trans | ||
| rs8022005 | chr14 | 61345380 | CCL3 | 6348 | 0.07 | trans | ||
| rs10148418 | chr14 | 67003872 | CCL3 | 6348 | 0.16 | trans | ||
| rs4905035 | chr14 | 93489262 | CCL3 | 6348 | 0.08 | trans | ||
| rs941539 | chr14 | 93561214 | CCL3 | 6348 | 3.076E-4 | trans | ||
| rs1885073 | chr14 | 104357181 | CCL3 | 6348 | 1.2E-5 | trans | ||
| rs16973929 | chr15 | 47395709 | CCL3 | 6348 | 0.11 | trans | ||
| rs8039808 | chr15 | 49939845 | CCL3 | 6348 | 0.18 | trans | ||
| rs2414703 | chr15 | 61904602 | CCL3 | 6348 | 0.15 | trans | ||
| rs289171 | chr15 | 62576053 | CCL3 | 6348 | 0.05 | trans | ||
| rs289161 | chr15 | 62589888 | CCL3 | 6348 | 0.13 | trans | ||
| rs2249982 | chr15 | 70236349 | CCL3 | 6348 | 3.615E-4 | trans | ||
| rs939043 | chr15 | 70236428 | CCL3 | 6348 | 3.336E-5 | trans | ||
| rs11858762 | chr15 | 81386594 | CCL3 | 6348 | 0.11 | trans | ||
| rs9635390 | chr15 | 90809632 | CCL3 | 6348 | 0.02 | trans | ||
| rs7174562 | chr15 | 93811530 | CCL3 | 6348 | 0.13 | trans | ||
| rs8047151 | chr16 | 17209696 | CCL3 | 6348 | 5.239E-5 | trans | ||
| rs4271578 | chr16 | 17213886 | CCL3 | 6348 | 4.583E-4 | trans | ||
| rs7200513 | chr16 | 17219474 | CCL3 | 6348 | 0.13 | trans | ||
| rs3829495 | chr16 | 17221986 | CCL3 | 6348 | 0.01 | trans | ||
| rs7193763 | chr16 | 17225534 | CCL3 | 6348 | 0 | trans | ||
| rs11861635 | chr16 | 82543377 | CCL3 | 6348 | 0.01 | trans | ||
| rs1867231 | chr17 | 13678834 | CCL3 | 6348 | 0.16 | trans | ||
| rs757075 | chr17 | 31372017 | CCL3 | 6348 | 0.17 | trans | ||
| rs7219986 | chr17 | 53263206 | CCL3 | 6348 | 0.08 | trans | ||
| rs17817797 | chr17 | 53332496 | CCL3 | 6348 | 5.712E-4 | trans | ||
| rs16951770 | chr18 | 7444826 | CCL3 | 6348 | 0.1 | trans | ||
| rs16951384 | chr18 | 47527116 | CCL3 | 6348 | 0.09 | trans | ||
| rs8095378 | chr18 | 49283781 | CCL3 | 6348 | 0.05 | trans | ||
| rs17074993 | chr18 | 63260128 | CCL3 | 6348 | 0.12 | trans | ||
| snp_a-1821751 | 0 | CCL3 | 6348 | 0.01 | trans | |||
| rs1942464 | chr18 | 70509877 | CCL3 | 6348 | 0.13 | trans | ||
| rs17056922 | chr18 | 73086804 | CCL3 | 6348 | 0.2 | trans | ||
| rs1540047 | chr18 | 75047028 | CCL3 | 6348 | 0 | trans | ||
| rs2850845 | chr18 | 75048150 | CCL3 | 6348 | 0.02 | trans | ||
| rs2850843 | chr18 | 75048905 | CCL3 | 6348 | 0 | trans | ||
| rs1540051 | chr18 | 75049934 | CCL3 | 6348 | 0 | trans | ||
| rs6603096 | chr19 | 7415722 | CCL3 | 6348 | 0.15 | trans | ||
| rs16980096 | chr19 | 15000921 | CCL3 | 6348 | 0.18 | trans | ||
| snp_a-2138575 | 0 | CCL3 | 6348 | 0.04 | trans | |||
| rs234363 | chr19 | 40474607 | CCL3 | 6348 | 0.19 | trans | ||
| rs6417634 | chr20 | 6408617 | CCL3 | 6348 | 0.2 | trans | ||
| rs16997215 | chr20 | 16101890 | CCL3 | 6348 | 0.04 | trans | ||
| rs6026122 | chr20 | 56769083 | CCL3 | 6348 | 0.1 | trans | ||
| rs6015452 | chr20 | 57782312 | CCL3 | 6348 | 0.16 | trans | ||
| rs6124034 | chr20 | 59517206 | CCL3 | 6348 | 0.11 | trans | ||
| rs6124036 | chr20 | 59517478 | CCL3 | 6348 | 0.11 | trans | ||
| rs4812237 | chr20 | 59518040 | CCL3 | 6348 | 0.03 | trans | ||
| rs6586271 | chr21 | 44362641 | CCL3 | 6348 | 0.1 | trans | ||
| rs134028 | chr22 | 28048448 | CCL3 | 6348 | 0.01 | trans | ||
| rs5762317 | chr22 | 28094988 | CCL3 | 6348 | 0.07 | trans | ||
| rs5768165 | chr22 | 48318197 | CCL3 | 6348 | 0.01 | trans | ||
| rs5768168 | chr22 | 48319334 | CCL3 | 6348 | 0.01 | trans | ||
| rs5768173 | chr22 | 48321475 | CCL3 | 6348 | 0.01 | trans | ||
| rs6567620 | 0 | CCL3 | 6348 | 0.14 | trans | |||
| rs12006821 | chrX | 50424687 | CCL3 | 6348 | 0.13 | trans | ||
| rs16992781 | chrX | 146460376 | CCL3 | 6348 | 0.05 | trans | ||
| rs4824295 | chrX | 146496881 | CCL3 | 6348 | 0.02 | trans |
Section II. Transcriptome annotation
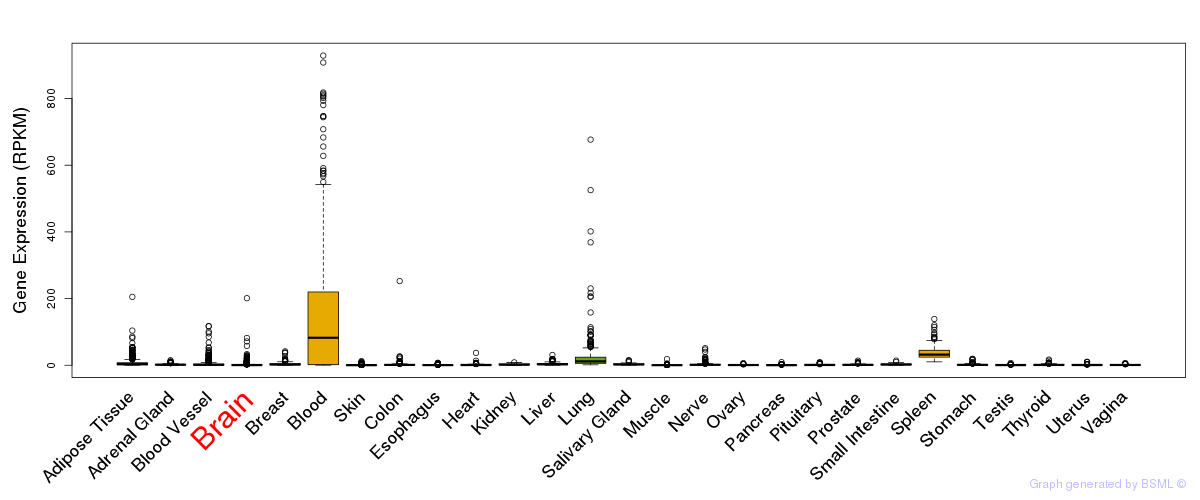
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004871 | signal transducer activity | TAS | 10857861 | |
| GO:0008009 | chemokine activity | TAS | 10660125 | |
| GO:0042056 | chemoattractant activity | IDA | 15001559 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007186 | G-protein coupled receptor protein signaling pathway | TAS | 10734056 | |
| GO:0007267 | cell-cell signaling | TAS | 10679098 | |
| GO:0007010 | cytoskeleton organization | TAS | 10072545 | |
| GO:0007165 | signal transduction | TAS | 10706735 | |
| GO:0006955 | immune response | IEA | - | |
| GO:0006954 | inflammatory response | TAS | 10679098 | |
| GO:0006935 | chemotaxis | TAS | 10679098 | |
| GO:0006928 | cell motion | TAS | 10660125 | |
| GO:0006887 | exocytosis | TAS | 10734056 | |
| GO:0006874 | cellular calcium ion homeostasis | TAS | 10734056 | |
| GO:0045069 | regulation of viral genome replication | TAS | 10841574 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005615 | extracellular space | IEA | - | |
| GO:0005625 | soluble fraction | TAS | 10706735 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BCL6 | BCL5 | BCL6A | LAZ3 | ZBTB27 | ZNF51 | B-cell CLL/lymphoma 6 | BCL6 interacts with the CCL3 promoter region. | BIND | 15531890 |
| CCBP2 | CCR10 | CCR9 | CMKBR9 | D6 | MGC126678 | MGC138250 | hD6 | chemokine binding protein 2 | - | HPRD | 9405404 |
| CCL3 | G0S19-1 | LD78ALPHA | MIP-1-alpha | MIP1A | SCYA3 | chemokine (C-C motif) ligand 3 | - | HPRD | 8460127 |
| CCL3 | G0S19-1 | LD78ALPHA | MIP-1-alpha | MIP1A | SCYA3 | chemokine (C-C motif) ligand 3 | Reconstituted Complex | BioGRID | 8077676 |
| CCL4 | ACT2 | AT744.1 | G-26 | LAG1 | MGC104418 | MGC126025 | MGC126026 | MIP-1-beta | MIP1B | MIP1B1 | SCYA2 | SCYA4 | chemokine (C-C motif) ligand 4 | - | HPRD,BioGRID | 11278300 |
| CCR1 | CD191 | CKR-1 | CKR1 | CMKBR1 | HM145 | MIP1aR | SCYAR1 | chemokine (C-C motif) receptor 1 | - | HPRD | 10202040|10660125 |
| CCR4 | CC-CKR-4 | CD194 | CKR4 | CMKBR4 | ChemR13 | HGCN:14099 | K5-5 | MGC88293 | chemokine (C-C motif) receptor 4 | - | HPRD,BioGRID | 8573157 |
| CCR5 | CC-CKR-5 | CCCKR5 | CD195 | CKR-5 | CKR5 | CMKBR5 | chemokine (C-C motif) receptor 5 | - | HPRD,BioGRID | 11700073 |
| NCOR1 | KIAA1047 | MGC104216 | N-CoR | TRAC1 | hCIT529I10 | hN-CoR | nuclear receptor co-repressor 1 | NcoR interacts with the CCL3 promoter region. | BIND | 15531890 |
| NCOR2 | CTG26 | SMRT | SMRTE | SMRTE-tau | TNRC14 | TRAC-1 | TRAC1 | nuclear receptor co-repressor 2 | SMRT interacts with the CCL3 promoter region. | BIND | 15531890 |
| SRGN | FLJ12930 | MGC9289 | PPG | PRG | PRG1 | serglycin | - | HPRD | 8613703 |
| TGFB1 | CED | DPD1 | TGFB | TGFbeta | transforming growth factor, beta 1 | - | HPRD | 8568261 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CYTOKINE CYTOKINE RECEPTOR INTERACTION | 267 | 161 | All SZGR 2.0 genes in this pathway |
| KEGG CHEMOKINE SIGNALING PATHWAY | 190 | 128 | All SZGR 2.0 genes in this pathway |
| KEGG TOLL LIKE RECEPTOR SIGNALING PATHWAY | 102 | 88 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ERYTH PATHWAY | 15 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NKT PATHWAY | 29 | 21 | All SZGR 2.0 genes in this pathway |
| PID IL12 2PATHWAY | 63 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME PEPTIDE LIGAND BINDING RECEPTORS | 188 | 108 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | 305 | 177 | All SZGR 2.0 genes in this pathway |
| REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | 57 | 27 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR LIGAND BINDING | 408 | 246 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 CHRONIC LOF DN | 118 | 78 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 ACUTE LOF UP | 215 | 137 | All SZGR 2.0 genes in this pathway |
| DEBOSSCHER NFKB TARGETS REPRESSED BY GLUCOCORTICOIDS | 24 | 18 | All SZGR 2.0 genes in this pathway |
| DIRMEIER LMP1 RESPONSE EARLY | 66 | 48 | All SZGR 2.0 genes in this pathway |
| FUNG IL2 SIGNALING 2 | 12 | 8 | All SZGR 2.0 genes in this pathway |
| FUNG IL2 TARGETS WITH STAT5 BINDING SITES T1 | 9 | 7 | All SZGR 2.0 genes in this pathway |
| TSAI RESPONSE TO IONIZING RADIATION | 149 | 101 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| SHIN B CELL LYMPHOMA CLUSTER 1 | 13 | 11 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPLICATION GENES | 147 | 87 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 17Q11 Q21 AMPLICON | 133 | 78 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| MAGRANGEAS MULTIPLE MYELOMA IGLL VS IGLK UP | 42 | 24 | All SZGR 2.0 genes in this pathway |
| NAKAJIMA EOSINOPHIL | 30 | 20 | All SZGR 2.0 genes in this pathway |
| GALINDO IMMUNE RESPONSE TO ENTEROTOXIN | 85 | 67 | All SZGR 2.0 genes in this pathway |
| PARK HSC MARKERS | 44 | 31 | All SZGR 2.0 genes in this pathway |
| NEMETH INFLAMMATORY RESPONSE LPS UP | 88 | 64 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND UP | 77 | 52 | All SZGR 2.0 genes in this pathway |
| BASSO CD40 SIGNALING UP | 101 | 76 | All SZGR 2.0 genes in this pathway |
| BRUNO HEMATOPOIESIS | 66 | 48 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY UP | 86 | 48 | All SZGR 2.0 genes in this pathway |
| KEEN RESPONSE TO ROSIGLITAZONE DN | 106 | 68 | All SZGR 2.0 genes in this pathway |
| ZHANG ANTIVIRAL RESPONSE TO RIBAVIRIN DN | 51 | 32 | All SZGR 2.0 genes in this pathway |
| SAFFORD T LYMPHOCYTE ANERGY | 87 | 54 | All SZGR 2.0 genes in this pathway |
| STEARMAN LUNG CANCER EARLY VS LATE DN | 61 | 39 | All SZGR 2.0 genes in this pathway |
| SARRIO EPITHELIAL MESENCHYMAL TRANSITION DN | 154 | 101 | All SZGR 2.0 genes in this pathway |
| MARZEC IL2 SIGNALING UP | 115 | 80 | All SZGR 2.0 genes in this pathway |
| GAURNIER PSMD4 TARGETS | 73 | 55 | All SZGR 2.0 genes in this pathway |
| AUJLA IL22 AND IL17A SIGNALING | 11 | 5 | All SZGR 2.0 genes in this pathway |
| QI PLASMACYTOMA UP | 259 | 185 | All SZGR 2.0 genes in this pathway |
| CROMER TUMORIGENESIS UP | 63 | 36 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS DN | 210 | 128 | All SZGR 2.0 genes in this pathway |
| SEKI INFLAMMATORY RESPONSE LPS UP | 77 | 56 | All SZGR 2.0 genes in this pathway |
| CHAUHAN RESPONSE TO METHOXYESTRADIOL DN | 102 | 65 | All SZGR 2.0 genes in this pathway |
| GOLDRATH ANTIGEN RESPONSE | 346 | 192 | All SZGR 2.0 genes in this pathway |
| LINDSTEDT DENDRITIC CELL MATURATION A | 67 | 52 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| SHAFFER IRF4 TARGETS IN MYELOMA VS MATURE B LYMPHOCYTE | 101 | 76 | All SZGR 2.0 genes in this pathway |
| SHAFFER IRF4 TARGETS IN PLASMA CELL VS MATURE B LYMPHOCYTE | 67 | 51 | All SZGR 2.0 genes in this pathway |
| SHAFFER IRF4 TARGETS IN ACTIVATED B LYMPHOCYTE | 81 | 66 | All SZGR 2.0 genes in this pathway |
| CROONQUIST STROMAL STIMULATION UP | 60 | 42 | All SZGR 2.0 genes in this pathway |
| CROONQUIST NRAS SIGNALING UP | 41 | 24 | All SZGR 2.0 genes in this pathway |
| KUROZUMI RESPONSE TO ONCOCYTIC VIRUS | 44 | 30 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S1 | 237 | 159 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| LI INDUCED T TO NATURAL KILLER UP | 307 | 182 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN | 841 | 431 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS UP | 217 | 131 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS GROUP2 | 60 | 38 | All SZGR 2.0 genes in this pathway |
| JUBAN TARGETS OF SPI1 AND FLI1 DN | 92 | 60 | All SZGR 2.0 genes in this pathway |
| ALTEMEIER RESPONSE TO LPS WITH MECHANICAL VENTILATION | 128 | 81 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
| NABA SECRETED FACTORS | 344 | 197 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |