Gene Page: BRAF
Summary ?
| GeneID | 673 |
| Symbol | BRAF |
| Synonyms | B-RAF1|BRAF1|NS7|RAFB1 |
| Description | B-Raf proto-oncogene, serine/threonine kinase |
| Reference | MIM:164757|HGNC:HGNC:1097|Ensembl:ENSG00000157764|HPRD:01264|Vega:OTTHUMG00000157457 |
| Gene type | protein-coding |
| Map location | 7q34 |
| Pascal p-value | 0.068 |
| Fetal beta | 0.15 |
| DMG | 1 (# studies) |
| Support | CANABINOID DOPAMINE SEROTONIN |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0286 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg08428486 | 7 | 140452281 | BRAF | 3.34E-4 | -0.363 | 0.041 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
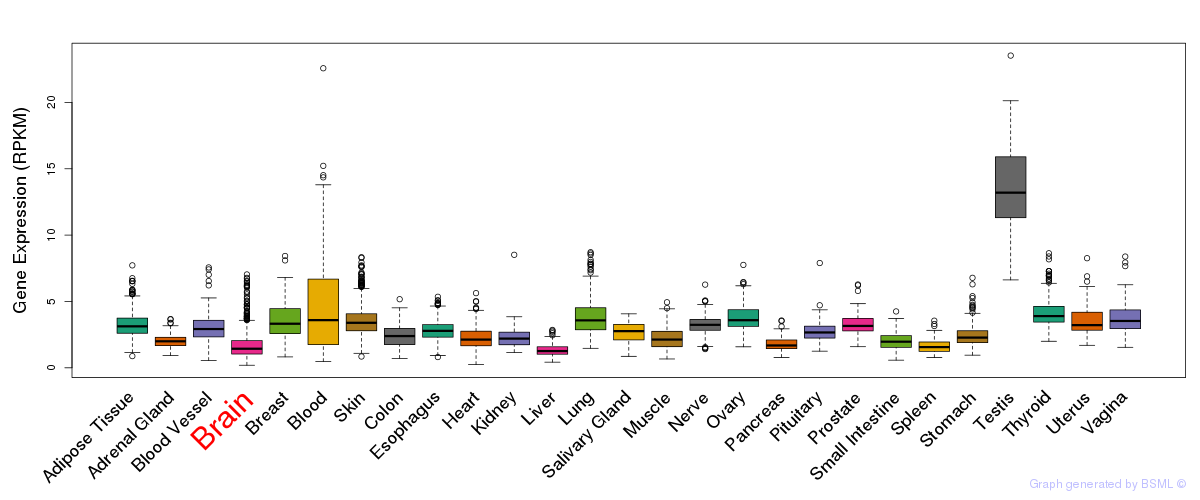
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ZFP36L2 | 0.85 | 0.87 |
| SOX9 | 0.83 | 0.73 |
| FBXL7 | 0.83 | 0.85 |
| SLC2A10 | 0.81 | 0.80 |
| PALLD | 0.79 | 0.75 |
| NR2E1 | 0.78 | 0.76 |
| CRB2 | 0.78 | 0.75 |
| YAP1 | 0.77 | 0.83 |
| NFATC4 | 0.77 | 0.71 |
| SIX5 | 0.77 | 0.64 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SYCP3 | -0.24 | -0.26 |
| ZBTB8OS | -0.22 | -0.28 |
| USMG5 | -0.21 | -0.30 |
| THOC7 | -0.21 | -0.25 |
| AF347015.21 | -0.21 | -0.15 |
| AL050337.1 | -0.21 | -0.27 |
| DDTL | -0.20 | -0.30 |
| BEX5 | -0.20 | -0.15 |
| GNRHR | -0.20 | -0.26 |
| FABP3 | -0.20 | -0.12 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005057 | receptor signaling protein activity | IEA | - | |
| GO:0005515 | protein binding | IPI | 17353931 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0004674 | protein serine/threonine kinase activity | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0019992 | diacylglycerol binding | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006468 | protein amino acid phosphorylation | TAS | 2284096 | |
| GO:0009887 | organ morphogenesis | TAS | 9207797 | |
| GO:0007264 | small GTPase mediated signal transduction | EXP | 10712923 | |
| GO:0006916 | anti-apoptosis | TAS | 9207797 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IDA | 18029348 | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AKT1 | AKT | MGC99656 | PKB | PKB-ALPHA | PRKBA | RAC | RAC-ALPHA | v-akt murine thymoma viral oncogene homolog 1 | - | HPRD,BioGRID | 10869359 |
| HRAS | C-BAS/HAS | C-H-RAS | C-HA-RAS1 | CTLO | H-RASIDX | HAMSV | HRAS1 | K-RAS | N-RAS | RASH1 | v-Ha-ras Harvey rat sarcoma viral oncogene homolog | - | HPRD,BioGRID | 7706312 |
| MAP2K1 | MAPKK1 | MEK1 | MKK1 | PRKMK1 | mitogen-activated protein kinase kinase 1 | MEK1 interacts with B-Raf. | BIND | 8621729 |
| MAP2K1 | MAPKK1 | MEK1 | MKK1 | PRKMK1 | mitogen-activated protein kinase kinase 1 | - | HPRD | 7731720 |
| MAP2K2 | FLJ26075 | MAPKK2 | MEK2 | MKK2 | PRKMK2 | mitogen-activated protein kinase kinase 2 | - | HPRD | 8668348 |
| MAPK3 | ERK1 | HS44KDAP | HUMKER1A | MGC20180 | P44ERK1 | P44MAPK | PRKM3 | mitogen-activated protein kinase 3 | Phenotypic Enhancement | BioGRID | 10869359 |
| MRAS | FLJ42964 | M-RAs | R-RAS3 | RRAS3 | muscle RAS oncogene homolog | - | HPRD,BioGRID | 12138204 |
| OIP5 | 5730547N13Rik | LINT-25 | MIS18beta | Opa interacting protein 5 | B-RAF interacts with OIP5. | BIND | 12620389 |
| RAF1 | CRAF | NS5 | Raf-1 | c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 | B-RAF interacts with C-RAF. This interaction was modelled on a demonstrated interaction between human B-RAF and monkey C-RAF . | BIND | 15035987 |
| RAF1 | CRAF | NS5 | Raf-1 | c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 | - | HPRD,BioGRID | 11325826 |
| RAP1A | KREV-1 | KREV1 | RAP1 | SMGP21 | RAP1A, member of RAS oncogene family | Reconstituted Complex | BioGRID | 10454553 |
| RAP1GAP | KIAA0474 | RAP1GA1 | Rap1GAP1 | rap1GAPII | RAP1 GTPase activating protein | Reconstituted Complex | BioGRID | 11278445 |
| SFN | YWHAS | stratifin | Affinity Capture-MS | BioGRID | 15778465 |
| TIMM50 | MGC102733 | TIM50 | TIM50L | translocase of inner mitochondrial membrane 50 homolog (S. cerevisiae) | B-RAF interacts with TIMM50. | BIND | 12620389 |
| YWHAB | GW128 | HS1 | KCIP-1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide | BRAF (B-Raf) interacts with an unspecified isoform of YWHAB (14-3-3-beta). | BIND | 12620389 |
| YWHAB | GW128 | HS1 | KCIP-1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide | B-RAF interacts with 14-3-3-beta. | BIND | 12620389 |
| YWHAB | GW128 | HS1 | KCIP-1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide | - | HPRD,BioGRID | 10931830 |
| YWHAG | 14-3-3GAMMA | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide | Affinity Capture-MS | BioGRID | 17353931 |
| YWHAH | YWHA1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide | - | HPRD | 8668348 |
| YWHAQ | 14-3-3 | 1C5 | HS1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide | - | HPRD | 8668348 |
| YWHAQ | 14-3-3 | 1C5 | HS1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide | Affinity Capture-MS | BioGRID | 17353931 |
| YWHAZ | KCIP-1 | MGC111427 | MGC126532 | MGC138156 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | Affinity Capture-MS | BioGRID | 17353931 |
| YWHAZ | KCIP-1 | MGC111427 | MGC126532 | MGC138156 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | - | HPRD | 8668348 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| KEGG ERBB SIGNALING PATHWAY | 87 | 71 | All SZGR 2.0 genes in this pathway |
| KEGG CHEMOKINE SIGNALING PATHWAY | 190 | 128 | All SZGR 2.0 genes in this pathway |
| KEGG MTOR SIGNALING PATHWAY | 52 | 40 | All SZGR 2.0 genes in this pathway |
| KEGG VASCULAR SMOOTH MUSCLE CONTRACTION | 115 | 81 | All SZGR 2.0 genes in this pathway |
| KEGG FOCAL ADHESION | 201 | 138 | All SZGR 2.0 genes in this pathway |
| KEGG NATURAL KILLER CELL MEDIATED CYTOTOXICITY | 137 | 92 | All SZGR 2.0 genes in this pathway |
| KEGG LONG TERM POTENTIATION | 70 | 57 | All SZGR 2.0 genes in this pathway |
| KEGG NEUROTROPHIN SIGNALING PATHWAY | 126 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG LONG TERM DEPRESSION | 70 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG REGULATION OF ACTIN CYTOSKELETON | 216 | 144 | All SZGR 2.0 genes in this pathway |
| KEGG INSULIN SIGNALING PATHWAY | 137 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG PROGESTERONE MEDIATED OOCYTE MATURATION | 86 | 59 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG COLORECTAL CANCER | 62 | 47 | All SZGR 2.0 genes in this pathway |
| KEGG RENAL CELL CARCINOMA | 70 | 60 | All SZGR 2.0 genes in this pathway |
| KEGG PANCREATIC CANCER | 70 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG ENDOMETRIAL CANCER | 52 | 45 | All SZGR 2.0 genes in this pathway |
| KEGG GLIOMA | 65 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG PROSTATE CANCER | 89 | 75 | All SZGR 2.0 genes in this pathway |
| KEGG THYROID CANCER | 29 | 26 | All SZGR 2.0 genes in this pathway |
| KEGG MELANOMA | 71 | 57 | All SZGR 2.0 genes in this pathway |
| KEGG BLADDER CANCER | 42 | 33 | All SZGR 2.0 genes in this pathway |
| KEGG CHRONIC MYELOID LEUKEMIA | 73 | 59 | All SZGR 2.0 genes in this pathway |
| KEGG ACUTE MYELOID LEUKEMIA | 60 | 47 | All SZGR 2.0 genes in this pathway |
| KEGG NON SMALL CELL LUNG CANCER | 54 | 47 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MAPK PATHWAY | 87 | 68 | All SZGR 2.0 genes in this pathway |
| ST DIFFERENTIATION PATHWAY IN PC12 CELLS | 45 | 35 | All SZGR 2.0 genes in this pathway |
| ST ERK1 ERK2 MAPK PATHWAY | 32 | 25 | All SZGR 2.0 genes in this pathway |
| ST G ALPHA S PATHWAY | 16 | 11 | All SZGR 2.0 genes in this pathway |
| ST G ALPHA I PATHWAY | 35 | 29 | All SZGR 2.0 genes in this pathway |
| ST ADRENERGIC | 36 | 29 | All SZGR 2.0 genes in this pathway |
| ST INTEGRIN SIGNALING PATHWAY | 82 | 62 | All SZGR 2.0 genes in this pathway |
| PID CDC42 PATHWAY | 70 | 51 | All SZGR 2.0 genes in this pathway |
| PID MTOR 4PATHWAY | 69 | 55 | All SZGR 2.0 genes in this pathway |
| PID TCR RAS PATHWAY | 14 | 14 | All SZGR 2.0 genes in this pathway |
| PID ERBB1 DOWNSTREAM PATHWAY | 105 | 78 | All SZGR 2.0 genes in this pathway |
| PID PDGFRB PATHWAY | 129 | 103 | All SZGR 2.0 genes in this pathway |
| PID VEGFR1 2 PATHWAY | 69 | 57 | All SZGR 2.0 genes in this pathway |
| PID MAPK TRK PATHWAY | 34 | 31 | All SZGR 2.0 genes in this pathway |
| PID CD8 TCR DOWNSTREAM PATHWAY | 65 | 56 | All SZGR 2.0 genes in this pathway |
| PID FAK PATHWAY | 59 | 45 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME SPRY REGULATION OF FGF SIGNALING | 14 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME NEGATIVE REGULATION OF FGFR SIGNALING | 37 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME ARMS MEDIATED ACTIVATION | 17 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME PROLONGED ERK ACTIVATION EVENTS | 19 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | 137 | 105 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING TO ERKS | 36 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR IN DISEASE | 127 | 88 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING TO P38 VIA RIT AND RIN | 15 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | 186 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| REACTOME NEUROTRANSMITTER RECEPTOR BINDING AND DOWNSTREAM TRANSMISSION IN THE POSTSYNAPTIC CELL | 137 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF NMDA RECEPTOR UPON GLUTAMATE BINDING AND POSTSYNAPTIC EVENTS | 37 | 32 | All SZGR 2.0 genes in this pathway |
| REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | 27 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | 33 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR | 112 | 80 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS UP | 473 | 314 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS DN | 411 | 249 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR UP | 240 | 152 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 16HR UP | 225 | 139 | All SZGR 2.0 genes in this pathway |
| SU TESTIS | 76 | 53 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS BY HNE AND TBH | 60 | 42 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 10HR UP | 101 | 69 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 12HR UP | 111 | 68 | All SZGR 2.0 genes in this pathway |
| LIN MELANOMA COPY NUMBER UP | 73 | 53 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D UP | 210 | 124 | All SZGR 2.0 genes in this pathway |
| FIRESTEIN PROLIFERATION | 175 | 125 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| PANGAS TUMOR SUPPRESSION BY SMAD1 AND SMAD5 DN | 157 | 106 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 UP | 344 | 215 | All SZGR 2.0 genes in this pathway |