Gene Page: STAT1
Summary ?
| GeneID | 6772 |
| Symbol | STAT1 |
| Synonyms | CANDF7|IMD31A|IMD31B|IMD31C|ISGF-3|STAT91 |
| Description | signal transducer and activator of transcription 1 |
| Reference | MIM:600555|HGNC:HGNC:11362|Ensembl:ENSG00000115415|HPRD:02777|Vega:OTTHUMG00000132699 |
| Gene type | protein-coding |
| Map location | 2q32.2 |
| Pascal p-value | 0.007 |
| Sherlock p-value | 0.982 |
| Fetal beta | -1.421 |
| eGene | Myers' cis & trans |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.humanPSD G2Cdb.humanPSP |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.164 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17097182 | chr1 | 65903444 | STAT1 | 6772 | 0.1 | trans | ||
| rs17092900 | chr1 | 73003409 | STAT1 | 6772 | 1.514E-6 | trans | ||
| rs17121311 | chr1 | 100314968 | STAT1 | 6772 | 0.2 | trans | ||
| rs7532142 | chr1 | 157362311 | STAT1 | 6772 | 0.08 | trans | ||
| rs6427374 | chr1 | 157362489 | STAT1 | 6772 | 0.08 | trans | ||
| rs10494571 | chr1 | 183930700 | STAT1 | 6772 | 0.06 | trans | ||
| rs16857588 | chr2 | 11675145 | STAT1 | 6772 | 0 | trans | ||
| snp_a-2142922 | 0 | STAT1 | 6772 | 0.04 | trans | |||
| rs1900263 | chr2 | 80376061 | STAT1 | 6772 | 0.01 | trans | ||
| rs10496239 | chr2 | 80376968 | STAT1 | 6772 | 0.01 | trans | ||
| rs11679783 | chr2 | 80377391 | STAT1 | 6772 | 0.01 | trans | ||
| rs17018721 | chr2 | 80385148 | STAT1 | 6772 | 0.03 | trans | ||
| rs2862022 | chr2 | 80388509 | STAT1 | 6772 | 0.01 | trans | ||
| rs17043176 | chr2 | 114025371 | STAT1 | 6772 | 0 | trans | ||
| rs17490240 | chr2 | 146573997 | STAT1 | 6772 | 0.06 | trans | ||
| rs842573 | chr2 | 184293146 | STAT1 | 6772 | 0.07 | trans | ||
| rs11897086 | chr2 | 184596267 | STAT1 | 6772 | 0.14 | trans | ||
| rs10187085 | chr2 | 184614775 | STAT1 | 6772 | 0.06 | trans | ||
| rs9989867 | chr2 | 184628012 | STAT1 | 6772 | 0.06 | trans | ||
| rs17033273 | chr3 | 10676696 | STAT1 | 6772 | 0.08 | trans | ||
| rs7637875 | chr3 | 40594263 | STAT1 | 6772 | 0.06 | trans | ||
| rs277655 | chr3 | 100044586 | STAT1 | 6772 | 0.02 | trans | ||
| rs890527 | chr3 | 140774852 | STAT1 | 6772 | 0.01 | trans | ||
| rs6789808 | chr3 | 154492082 | STAT1 | 6772 | 0.09 | trans | ||
| rs7612673 | chr3 | 154499328 | STAT1 | 6772 | 3.968E-4 | trans | ||
| rs17502924 | chr4 | 16968009 | STAT1 | 6772 | 0.07 | trans | ||
| rs13110653 | chr4 | 16973010 | STAT1 | 6772 | 0.09 | trans | ||
| rs10461085 | chr4 | 30717743 | STAT1 | 6772 | 0.18 | trans | ||
| rs11942278 | chr4 | 58922687 | STAT1 | 6772 | 0.05 | trans | ||
| rs6551878 | chr4 | 65690588 | STAT1 | 6772 | 0.16 | trans | ||
| rs7688236 | chr4 | 86804077 | STAT1 | 6772 | 0.17 | trans | ||
| rs2653828 | chr4 | 173851087 | STAT1 | 6772 | 0.03 | trans | ||
| rs159171 | chr5 | 21186325 | STAT1 | 6772 | 0.13 | trans | ||
| rs1678893 | chr5 | 31763244 | STAT1 | 6772 | 1.603E-7 | trans | ||
| rs10515260 | chr5 | 97076547 | STAT1 | 6772 | 0.02 | trans | ||
| rs4705861 | chr5 | 131800763 | STAT1 | 6772 | 0.09 | trans | ||
| rs17101114 | chr5 | 143246001 | STAT1 | 6772 | 0.18 | trans | ||
| rs10516136 | chr5 | 176381368 | STAT1 | 6772 | 0.04 | trans | ||
| rs13176869 | chr5 | 179203478 | STAT1 | 6772 | 6.028E-5 | trans | ||
| rs9396602 | chr6 | 15766500 | STAT1 | 6772 | 0.18 | trans | ||
| rs1240825 | chr6 | 16862984 | STAT1 | 6772 | 0.08 | trans | ||
| rs7741955 | chr6 | 84829621 | STAT1 | 6772 | 0.08 | trans | ||
| rs12524136 | chr6 | 84935440 | STAT1 | 6772 | 2.815E-4 | trans | ||
| rs16883826 | chr6 | 91671158 | STAT1 | 6772 | 0.01 | trans | ||
| rs9397692 | chr6 | 154487314 | STAT1 | 6772 | 7.187E-6 | trans | ||
| rs10485060 | chr6 | 154516100 | STAT1 | 6772 | 9.442E-6 | trans | ||
| rs9384190 | chr6 | 154524473 | STAT1 | 6772 | 1.354E-5 | trans | ||
| rs17292544 | chr6 | 154544809 | STAT1 | 6772 | 1.017E-7 | trans | ||
| rs6462122 | chr7 | 29137630 | STAT1 | 6772 | 0.12 | trans | ||
| rs17171716 | chr7 | 40505332 | STAT1 | 6772 | 0.01 | trans | ||
| rs17160749 | chr7 | 77689922 | STAT1 | 6772 | 0.01 | trans | ||
| rs3779321 | chr7 | 77691378 | STAT1 | 6772 | 0.01 | trans | ||
| rs11974883 | chr7 | 78068523 | STAT1 | 6772 | 0.01 | trans | ||
| rs7807249 | chr7 | 131615735 | STAT1 | 6772 | 0.08 | trans | ||
| rs1355060 | chr8 | 118717710 | STAT1 | 6772 | 0.1 | trans | ||
| rs2280828 | chr8 | 118762435 | STAT1 | 6772 | 0.05 | trans | ||
| rs10283612 | chr9 | 1830285 | STAT1 | 6772 | 0.01 | trans | ||
| rs10963533 | chr9 | 1832589 | STAT1 | 6772 | 0.05 | trans | ||
| rs10283729 | chr9 | 2981636 | STAT1 | 6772 | 0.01 | trans | ||
| rs10816182 | chr9 | 9795184 | STAT1 | 6772 | 0.17 | trans | ||
| rs10752351 | chr10 | 6588016 | STAT1 | 6772 | 0.12 | trans | ||
| rs10796268 | chr10 | 6603916 | STAT1 | 6772 | 0.12 | trans | ||
| rs9416364 | chr10 | 56253021 | STAT1 | 6772 | 0.11 | trans | ||
| rs16916549 | chr10 | 63495204 | STAT1 | 6772 | 0.05 | trans | ||
| rs17111364 | chr10 | 97901813 | STAT1 | 6772 | 0.01 | trans | ||
| rs894376 | chr10 | 99674122 | STAT1 | 6772 | 0.08 | trans | ||
| rs10833144 | chr11 | 19656451 | STAT1 | 6772 | 0.03 | trans | ||
| rs11038039 | chr11 | 44530644 | STAT1 | 6772 | 0.04 | trans | ||
| rs11038047 | chr11 | 44543848 | STAT1 | 6772 | 0.02 | trans | ||
| rs2508654 | chr11 | 120405519 | STAT1 | 6772 | 0.16 | trans | ||
| rs17220663 | chr12 | 13859583 | STAT1 | 6772 | 0.12 | trans | ||
| rs2605422 | chr12 | 59148638 | STAT1 | 6772 | 0.15 | trans | ||
| rs2645865 | chr12 | 59151393 | STAT1 | 6772 | 0.17 | trans | ||
| rs2712516 | chr12 | 59152364 | STAT1 | 6772 | 0.07 | trans | ||
| rs3751152 | chr12 | 121442063 | STAT1 | 6772 | 0.03 | trans | ||
| rs9578342 | 0 | STAT1 | 6772 | 0.1 | trans | |||
| rs4769240 | chr13 | 23697920 | STAT1 | 6772 | 0.15 | trans | ||
| rs17061055 | chr13 | 40835399 | STAT1 | 6772 | 2.821E-7 | trans | ||
| rs17066364 | chr13 | 45515988 | STAT1 | 6772 | 1.066E-8 | trans | ||
| rs3783149 | chr13 | 45582764 | STAT1 | 6772 | 2.319E-4 | trans | ||
| rs7332969 | chr13 | 45612902 | STAT1 | 6772 | 1.066E-8 | trans | ||
| rs10484008 | chr14 | 89376562 | STAT1 | 6772 | 0.1 | trans | ||
| rs10150770 | chr14 | 95375311 | STAT1 | 6772 | 0.09 | trans | ||
| rs16955413 | chr15 | 29677652 | STAT1 | 6772 | 1.914E-7 | trans | ||
| rs4457950 | chr15 | 35445709 | STAT1 | 6772 | 0.08 | trans | ||
| rs16969948 | chr15 | 78864785 | STAT1 | 6772 | 0.03 | trans | ||
| rs6497089 | chr15 | 94163348 | STAT1 | 6772 | 0.16 | trans | ||
| rs3794689 | chr16 | 71600536 | STAT1 | 6772 | 0.09 | trans | ||
| rs963303 | chr16 | 73372497 | STAT1 | 6772 | 0.05 | trans | ||
| rs16955238 | chr16 | 81415690 | STAT1 | 6772 | 1.407E-4 | trans | ||
| rs16955255 | chr16 | 81420130 | STAT1 | 6772 | 4.711E-5 | trans | ||
| rs16955259 | chr16 | 81420188 | STAT1 | 6772 | 1.407E-4 | trans | ||
| rs416841 | chr16 | 86774391 | STAT1 | 6772 | 0.14 | trans | ||
| rs4303487 | chr16 | 90033254 | STAT1 | 6772 | 0.01 | trans | ||
| rs8098388 | chr18 | 24080746 | STAT1 | 6772 | 0.1 | trans | ||
| rs8085520 | chr18 | 40184498 | STAT1 | 6772 | 0.13 | trans | ||
| rs16976379 | chr18 | 40198836 | STAT1 | 6772 | 0.16 | trans | ||
| rs8105452 | chr19 | 11854016 | STAT1 | 6772 | 0.03 | trans | ||
| rs8104298 | chr19 | 11867512 | STAT1 | 6772 | 9.207E-4 | trans | ||
| rs16979927 | chr20 | 55019249 | STAT1 | 6772 | 2.166E-9 | trans | ||
| rs2832206 | chr21 | 30504653 | STAT1 | 6772 | 0.03 | trans | ||
| rs137907 | chr22 | 50486591 | STAT1 | 6772 | 0.13 | trans | ||
| rs17331284 | chrX | 6962964 | STAT1 | 6772 | 0.04 | trans | ||
| rs17347454 | chrX | 23347700 | STAT1 | 6772 | 4.925E-9 | trans | ||
| rs5952435 | chrX | 47321870 | STAT1 | 6772 | 0.01 | trans | ||
| rs689365 | chrX | 97074769 | STAT1 | 6772 | 4.056E-5 | trans | ||
| rs2805901 | chrX | 138809001 | STAT1 | 6772 | 0.02 | trans | ||
| rs5953621 | chrX | 138832928 | STAT1 | 6772 | 0.02 | trans | ||
| rs1415887 | chrX | 138848048 | STAT1 | 6772 | 0.02 | trans | ||
| rs6528651 | chrX | 138878265 | STAT1 | 6772 | 0.02 | trans | ||
| rs2485732 | chrX | 138907985 | STAT1 | 6772 | 0.02 | trans | ||
| rs2491006 | chrX | 138916494 | STAT1 | 6772 | 0.02 | trans | ||
| rs17328709 | chrX | 138930939 | STAT1 | 6772 | 0.02 | trans | ||
| rs2491009 | chrX | 138944850 | STAT1 | 6772 | 0.02 | trans | ||
| rs4824924 | chrX | 139019135 | STAT1 | 6772 | 0.02 | trans | ||
| rs10856133 | 0 | STAT1 | 6772 | 6.683E-5 | trans | |||
| rs16995164 | chrX | 148536507 | STAT1 | 6772 | 0.03 | trans |
Section II. Transcriptome annotation
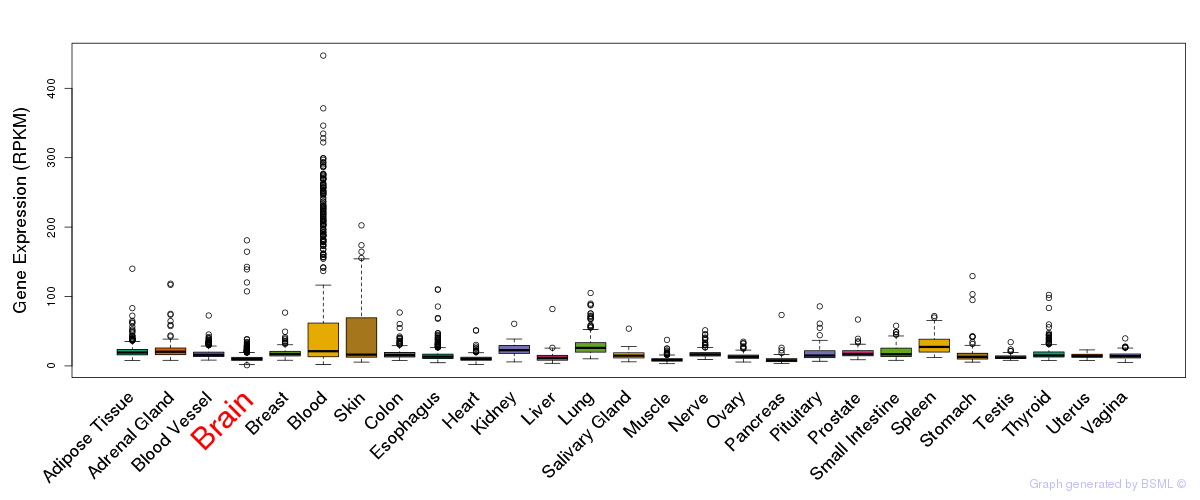
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AMIGO1 | 0.95 | 0.94 |
| ATP2B3 | 0.94 | 0.95 |
| SCAMP5 | 0.93 | 0.92 |
| CLSTN1 | 0.93 | 0.95 |
| MADD | 0.93 | 0.95 |
| STX1B | 0.93 | 0.91 |
| PIP4K2C | 0.92 | 0.91 |
| KIAA0513 | 0.92 | 0.92 |
| PI4KA | 0.92 | 0.93 |
| ABR | 0.91 | 0.93 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| DBI | -0.55 | -0.63 |
| RAB34 | -0.53 | -0.60 |
| GNG11 | -0.52 | -0.57 |
| AP002478.3 | -0.51 | -0.56 |
| RAB13 | -0.51 | -0.60 |
| C1orf61 | -0.51 | -0.63 |
| C1orf54 | -0.50 | -0.58 |
| AF347015.21 | -0.50 | -0.48 |
| RHOC | -0.48 | -0.56 |
| ACSF2 | -0.48 | -0.50 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004871 | signal transducer activity | IEA | - | |
| GO:0003700 | transcription factor activity | IEA | - | |
| GO:0003700 | transcription factor activity | TAS | 10820245 |10848577 | |
| GO:0005062 | hematopoietin/interferon-class (D200-domain) cytokine receptor signal transducer activity | TAS | 8608597 | |
| GO:0005509 | calcium ion binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 16306601 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006366 | transcription from RNA polymerase II promoter | TAS | 9630226 | |
| GO:0007260 | tyrosine phosphorylation of STAT protein | TAS | 10692450 | |
| GO:0007165 | signal transduction | IEA | - | |
| GO:0007165 | signal transduction | TAS | 10973496 | |
| GO:0009615 | response to virus | IEA | - | |
| GO:0006919 | caspase activation | TAS | 10692450 | |
| GO:0007249 | I-kappaB kinase/NF-kappaB cascade | TAS | 10848577 | |
| GO:0044419 | interspecies interaction between organisms | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IDA | 16306601 | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005730 | nucleolus | IDA | 18029348 | |
| GO:0005730 | nucleolus | IDA | 18029348 | |
| GO:0005737 | cytoplasm | TAS | 10820245 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ADRA1B | ADRA1 | ALPHA1BAR | adrenergic, alpha-1B-, receptor | - | HPRD | 10652206 |
| BMX | ETK | PSCTK2 | PSCTK3 | BMX non-receptor tyrosine kinase | - | HPRD | 9373245 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | - | HPRD,BioGRID | 10792030 |
| C20orf185 | LPLUNC3 | RYA3 | dJ726C3.4 | chromosome 20 open reading frame 185 | Affinity Capture-MS | BioGRID | 17353931 |
| CAMK2G | CAMK | CAMK-II | CAMKG | FLJ16043 | MGC26678 | calcium/calmodulin-dependent protein kinase II gamma | - | HPRD | 11972023 |
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | Stat1-beta interacts with CBP. | BIND | 8986769 |
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | Stat1-alpha interacts with CBP. | BIND | 8986769 |
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | - | HPRD,BioGRID | 8986769 |
| CSE1L | CAS | CSE1 | MGC117283 | MGC130036 | MGC130037 | XPO2 | CSE1 chromosome segregation 1-like (yeast) | - | HPRD | 11927559 |
| EGFR | ERBB | ERBB1 | HER1 | PIG61 | mENA | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | - | HPRD,BioGRID | 12070153 |
| EIF1AD | MGC11102 | eukaryotic translation initiation factor 1A domain containing | Two-hybrid | BioGRID | 16189514 |
| EIF2AK2 | EIF2AK1 | MGC126524 | PKR | PRKR | eukaryotic translation initiation factor 2-alpha kinase 2 | - | HPRD,BioGRID | 9135145 |11278865 |
| ELP2 | FLJ10879 | SHINC-2 | STATIP1 | StIP | elongation protein 2 homolog (S. cerevisiae) | - | HPRD,BioGRID | 10954736 |
| EP300 | KAT3B | p300 | E1A binding protein p300 | Affinity Capture-Western | BioGRID | 10464260 |
| EP300 | KAT3B | p300 | E1A binding protein p300 | Stat1-beta interacts with p300. This interaction is modelled on a demonstrated interaction between human Stat1-beta and p300 from an unspecified species. | BIND | 8986769 |
| FANCC | FA3 | FAC | FACC | FLJ14675 | Fanconi anemia, complementation group C | - | HPRD,BioGRID | 10848598 |
| FRAP1 | FLJ44809 | FRAP | FRAP2 | MTOR | RAFT1 | RAPT1 | FK506 binding protein 12-rapamycin associated protein 1 | Affinity Capture-Western | BioGRID | 12807916 |
| GNB2L1 | Gnb2-rs1 | H12.3 | HLC-7 | PIG21 | RACK1 | guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 | - | HPRD,BioGRID | 11301323 |
| GNB2L1 | Gnb2-rs1 | H12.3 | HLC-7 | PIG21 | RACK1 | guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 | RACK-1 interacts with STAT1-beta. | BIND | 12960323 |
| GNB2L1 | Gnb2-rs1 | H12.3 | HLC-7 | PIG21 | RACK1 | guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 | RACK-1 interacts with STAT1-alpha. | BIND | 12960323 |
| GTF2I | BAP-135 | BAP135 | BTKAP1 | DIWS | FLJ38776 | FLJ56355 | IB291 | SPIN | TFII-I | WBS | WBSCR6 | general transcription factor II, i | - | HPRD,BioGRID | 9584171 |
| HSF1 | HSTF1 | heat shock transcription factor 1 | - | HPRD,BioGRID | 9880553 |
| IFNAR2 | IFN-R | IFN-alpha-REC | IFNABR | IFNARB | interferon (alpha, beta and omega) receptor 2 | Reconstituted Complex | BioGRID | 9121453 |11301323 |
| IL27RA | CRL1 | IL27R | TCCR | WSX1 | zcytor1 | interleukin 27 receptor, alpha | - | HPRD,BioGRID | 12734330 |
| IL2RB | CD122 | P70-75 | interleukin 2 receptor, beta | - | HPRD,BioGRID | 10602027 |
| IL2RG | CD132 | IMD4 | SCIDX | SCIDX1 | interleukin 2 receptor, gamma (severe combined immunodeficiency) | - | HPRD | 11418623 |
| IRF1 | IRF-1 | MAR | interferon regulatory factor 1 | - | HPRD,BioGRID | 10764778 |
| IRF9 | IRF-9 | ISGF3 | ISGF3G | p48 | interferon regulatory factor 9 | - | HPRD,BioGRID | 8943351 |
| JAK1 | JAK1A | JAK1B | JTK3 | Janus kinase 1 (a protein tyrosine kinase) | - | HPRD | 11722592 |
| JUN | AP-1 | AP1 | c-Jun | jun oncogene | - | HPRD,BioGRID | 10490649 |
| KIT | C-Kit | CD117 | PBT | SCFR | v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | STAT1-alpha interacts with c-kit | BIND | 9355737 |
| KIT | C-Kit | CD117 | PBT | SCFR | v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | - | HPRD,BioGRID | 9355737 |
| KPNA1 | IPOA5 | NPI-1 | RCH2 | SRP1 | karyopherin alpha 1 (importin alpha 5) | - | HPRD,BioGRID | 12048190 |
| KPNA2 | IPOA1 | QIP2 | RCH1 | SRP1alpha | karyopherin alpha 2 (RAG cohort 1, importin alpha 1) | An unspecified isoform of STAT1 interacts with karyopherin alpha (K-alpha). | BIND | 10964507 |
| LMO2 | RBTN2 | RBTNL1 | RHOM2 | TTG2 | LIM domain only 2 (rhombotin-like 1) | Two-hybrid | BioGRID | 16189514 |
| MCM3 | HCC5 | MGC1157 | P1-MCM3 | P1.h | RLFB | minichromosome maintenance complex component 3 | - | HPRD | 11248027 |
| MCM5 | CDC46 | MGC5315 | P1-CDC46 | minichromosome maintenance complex component 5 | - | HPRD,BioGRID | 9843502 |11248027 |
| MDK | FLJ27379 | MK | NEGF2 | midkine (neurite growth-promoting factor 2) | - | HPRD | 9452495 |
| NMI | - | N-myc (and STAT) interactor | - | HPRD,BioGRID | 9989503 |
| NMI | - | N-myc (and STAT) interactor | Nmi interacts with Stat1 | BIND | 9989503 |
| PIAS1 | DDXBP1 | GBP | GU/RH-II | MGC141878 | MGC141879 | ZMIZ3 | protein inhibitor of activated STAT, 1 | - | HPRD,BioGRID | 9724754 |10805787 |12855578|10805787 |
| PIAS1 | DDXBP1 | GBP | GU/RH-II | MGC141878 | MGC141879 | ZMIZ3 | protein inhibitor of activated STAT, 1 | - | HPRD | 9724754 |10805787 |12855578 |
| PRKCD | MAY1 | MGC49908 | PKCD | nPKC-delta | protein kinase C, delta | Affinity Capture-Western | BioGRID | 12807916 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | - | HPRD,BioGRID | 11278462 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | - | HPRD,BioGRID | 9344858 |
| STAT1 | DKFZp686B04100 | ISGF-3 | STAT91 | signal transducer and activator of transcription 1, 91kDa | - | HPRD,BioGRID | 9630226 |
| STAT1 | DKFZp686B04100 | ISGF-3 | STAT91 | signal transducer and activator of transcription 1, 91kDa | Stat1-alpha interacts with Stat1-alpha. This interaction was modelled on a demonstrated interaction between Stat1-alpha and Stat1-alpha both from an unspecified species. | BIND | 8986769 |
| STAT1 | DKFZp686B04100 | ISGF-3 | STAT91 | signal transducer and activator of transcription 1, 91kDa | Stat1 interacts with itself to form a dimer. | BIND | 15780933 |
| STAT2 | ISGF-3 | MGC59816 | P113 | STAT113 | signal transducer and activator of transcription 2, 113kDa | STAT1 interacts with STAT2. | BIND | 15467722 |
| STAT2 | ISGF-3 | MGC59816 | P113 | STAT113 | signal transducer and activator of transcription 2, 113kDa | - | HPRD,BioGRID | 8621447 |10446176 |
| STAT3 | APRF | FLJ20882 | HIES | MGC16063 | signal transducer and activator of transcription 3 (acute-phase response factor) | - | HPRD,BioGRID | 12070153 |
| SYK | DKFZp313N1010 | FLJ25043 | FLJ37489 | spleen tyrosine kinase | Syk interacts with and phosphorylates STAT1-alpha. | BIND | 15467722 |
| SYK | DKFZp313N1010 | FLJ25043 | FLJ37489 | spleen tyrosine kinase | Syk interacts with STAT1-beta. | BIND | 15467722 |
| TNFRSF1A | CD120a | FPF | MGC19588 | TBP1 | TNF-R | TNF-R-I | TNF-R55 | TNFAR | TNFR1 | TNFR55 | TNFR60 | p55 | p55-R | p60 | tumor necrosis factor receptor superfamily, member 1A | - | HPRD,BioGRID | 10848577 |
| TNFRSF1B | CD120b | TBPII | TNF-R-II | TNF-R75 | TNFBR | TNFR1B | TNFR2 | TNFR80 | p75 | p75TNFR | tumor necrosis factor receptor superfamily, member 1B | - | HPRD | 10848577 |
| TRADD | Hs.89862 | MGC11078 | TNFRSF1A-associated via death domain | - | HPRD,BioGRID | 10848577 |
| TYK2 | JTK1 | tyrosine kinase 2 | Biochemical Activity | BioGRID | 12960323 |
| UBE2I | C358B7.1 | P18 | UBC9 | ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) | - | HPRD | 12356736 |
| VDR | NR1I1 | vitamin D (1,25- dihydroxyvitamin D3) receptor | - | HPRD,BioGRID | 11909970 |
| XPO1 | CRM1 | DKFZp686B1823 | exportin 1 (CRM1 homolog, yeast) | - | HPRD | 9205132 |
| ZNF467 | EZI | Zfp467 | zinc finger protein 467 | - | HPRD | 12426389 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CHEMOKINE SIGNALING PATHWAY | 190 | 128 | All SZGR 2.0 genes in this pathway |
| KEGG TOLL LIKE RECEPTOR SIGNALING PATHWAY | 102 | 88 | All SZGR 2.0 genes in this pathway |
| KEGG JAK STAT SIGNALING PATHWAY | 155 | 105 | All SZGR 2.0 genes in this pathway |
| KEGG LEISHMANIA INFECTION | 72 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG PANCREATIC CANCER | 70 | 56 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CHEMICAL PATHWAY | 22 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BIOPEPTIDES PATHWAY | 45 | 35 | All SZGR 2.0 genes in this pathway |
| BIOCARTA EGF PATHWAY | 31 | 25 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IL10 PATHWAY | 17 | 14 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IL22BP PATHWAY | 16 | 12 | All SZGR 2.0 genes in this pathway |
| BIOCARTA GLEEVEC PATHWAY | 23 | 17 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MAPK PATHWAY | 87 | 68 | All SZGR 2.0 genes in this pathway |
| BIOCARTA P38MAPK PATHWAY | 40 | 31 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PDGF PATHWAY | 32 | 25 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TPO PATHWAY | 24 | 17 | All SZGR 2.0 genes in this pathway |
| ST INTERFERON GAMMA PATHWAY | 10 | 7 | All SZGR 2.0 genes in this pathway |
| ST TYPE I INTERFERON PATHWAY | 9 | 8 | All SZGR 2.0 genes in this pathway |
| PID GMCSF PATHWAY | 37 | 31 | All SZGR 2.0 genes in this pathway |
| PID IL27 PATHWAY | 26 | 20 | All SZGR 2.0 genes in this pathway |
| PID IL12 2PATHWAY | 63 | 54 | All SZGR 2.0 genes in this pathway |
| PID TCPTP PATHWAY | 43 | 33 | All SZGR 2.0 genes in this pathway |
| PID SHP2 PATHWAY | 58 | 46 | All SZGR 2.0 genes in this pathway |
| PID REG GR PATHWAY | 82 | 60 | All SZGR 2.0 genes in this pathway |
| PID IL2 1PATHWAY | 55 | 43 | All SZGR 2.0 genes in this pathway |
| PID CXCR4 PATHWAY | 102 | 78 | All SZGR 2.0 genes in this pathway |
| PID ERBB1 RECEPTOR PROXIMAL PATHWAY | 35 | 29 | All SZGR 2.0 genes in this pathway |
| PID TNF PATHWAY | 46 | 33 | All SZGR 2.0 genes in this pathway |
| PID IFNG PATHWAY | 40 | 34 | All SZGR 2.0 genes in this pathway |
| PID ERBB1 DOWNSTREAM PATHWAY | 105 | 78 | All SZGR 2.0 genes in this pathway |
| PID IL6 7 PATHWAY | 47 | 40 | All SZGR 2.0 genes in this pathway |
| PID PDGFRB PATHWAY | 129 | 103 | All SZGR 2.0 genes in this pathway |
| PID IL23 PATHWAY | 37 | 30 | All SZGR 2.0 genes in this pathway |
| PID KIT PATHWAY | 52 | 40 | All SZGR 2.0 genes in this pathway |
| PID EPO PATHWAY | 34 | 29 | All SZGR 2.0 genes in this pathway |
| PID FGF PATHWAY | 55 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY SCF KIT | 78 | 59 | All SZGR 2.0 genes in this pathway |
| REACTOME GROWTH HORMONE RECEPTOR SIGNALING | 24 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | 66 | 45 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR IN DISEASE | 127 | 88 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR1 MUTANTS | 30 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | 19 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR MUTANTS | 44 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY PDGF | 122 | 93 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | 95 | 76 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ILS | 107 | 86 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF IFNG SIGNALING | 14 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME INTERFERON GAMMA SIGNALING | 63 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME INTERFERON ALPHA BETA SIGNALING | 64 | 50 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF IFNA SIGNALING | 24 | 22 | All SZGR 2.0 genes in this pathway |
| REACTOME INTERFERON SIGNALING | 159 | 116 | All SZGR 2.0 genes in this pathway |
| REACTOME IL 6 SIGNALING | 11 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM | 270 | 204 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 DN | 267 | 178 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| BERTUCCI MEDULLARY VS DUCTAL BREAST CANCER UP | 206 | 111 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA UP | 294 | 178 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS UP | 473 | 314 | All SZGR 2.0 genes in this pathway |
| SAMOLS TARGETS OF KHSV MIRNAS DN | 62 | 35 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE UP | 423 | 283 | All SZGR 2.0 genes in this pathway |
| NOJIMA SFRP2 TARGETS DN | 25 | 18 | All SZGR 2.0 genes in this pathway |
| WANG CLIM2 TARGETS DN | 186 | 114 | All SZGR 2.0 genes in this pathway |
| GARGALOVIC RESPONSE TO OXIDIZED PHOSPHOLIPIDS GREY DN | 74 | 44 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 3D UP | 182 | 110 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 8D UP | 157 | 91 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 10D UP | 194 | 122 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 16D UP | 175 | 108 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION ERYTHROCYTE UP | 157 | 104 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| HAHTOLA SEZARY SYNDROM UP | 98 | 58 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES UP | 19 | 15 | All SZGR 2.0 genes in this pathway |
| YEMELYANOV GR TARGETS DN | 10 | 8 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| CASTELLANO NRAS TARGETS UP | 68 | 41 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 ACUTE LOF UP | 215 | 137 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND SERUM DEPRIVATION UP | 211 | 136 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN UP | 612 | 367 | All SZGR 2.0 genes in this pathway |
| MISSIAGLIA REGULATED BY METHYLATION UP | 126 | 78 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| NUNODA RESPONSE TO DASATINIB IMATINIB DN | 13 | 10 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION | 77 | 51 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 TTD UP | 64 | 39 | All SZGR 2.0 genes in this pathway |
| SEIDEN ONCOGENESIS BY MET | 88 | 53 | All SZGR 2.0 genes in this pathway |
| LI AMPLIFIED IN LUNG CANCER | 178 | 108 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| PATTERSON DOCETAXEL RESISTANCE | 29 | 20 | All SZGR 2.0 genes in this pathway |
| BOWIE RESPONSE TO EXTRACELLULAR MATRIX | 17 | 11 | All SZGR 2.0 genes in this pathway |
| BOWIE RESPONSE TO TAMOXIFEN | 18 | 10 | All SZGR 2.0 genes in this pathway |
| EINAV INTERFERON SIGNATURE IN CANCER | 27 | 16 | All SZGR 2.0 genes in this pathway |
| DAUER STAT3 TARGETS DN | 50 | 34 | All SZGR 2.0 genes in this pathway |
| MEINHOLD OVARIAN CANCER LOW GRADE DN | 20 | 16 | All SZGR 2.0 genes in this pathway |
| YAN ESCAPE FROM ANOIKIS | 24 | 19 | All SZGR 2.0 genes in this pathway |
| WOOD EBV EBNA1 TARGETS UP | 110 | 71 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| TSAI RESPONSE TO IONIZING RADIATION | 149 | 101 | All SZGR 2.0 genes in this pathway |
| PUJANA BREAST CANCER LIT INT NETWORK | 101 | 73 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| KHETCHOUMIAN TRIM24 TARGETS UP | 47 | 38 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| FRIDMAN SENESCENCE UP | 77 | 60 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES CORE NINE CORRELATED | 100 | 68 | All SZGR 2.0 genes in this pathway |
| SAKAI CHRONIC HEPATITIS VS LIVER CANCER DN | 36 | 24 | All SZGR 2.0 genes in this pathway |
| SHIN B CELL LYMPHOMA CLUSTER 8 | 36 | 28 | All SZGR 2.0 genes in this pathway |
| SHIPP DLBCL CURED VS FATAL DN | 45 | 30 | All SZGR 2.0 genes in this pathway |
| GRANDVAUX IFN RESPONSE NOT VIA IRF3 | 14 | 10 | All SZGR 2.0 genes in this pathway |
| KIM GERMINAL CENTER T HELPER UP | 66 | 42 | All SZGR 2.0 genes in this pathway |
| JECHLINGER EPITHELIAL TO MESENCHYMAL TRANSITION UP | 71 | 51 | All SZGR 2.0 genes in this pathway |
| DER IFN ALPHA RESPONSE UP | 74 | 48 | All SZGR 2.0 genes in this pathway |
| IIZUKA LIVER CANCER PROGRESSION G1 G2 UP | 12 | 9 | All SZGR 2.0 genes in this pathway |
| RADAEVA RESPONSE TO IFNA1 UP | 52 | 40 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK UP | 87 | 58 | All SZGR 2.0 genes in this pathway |
| WIELAND UP BY HBV INFECTION | 101 | 66 | All SZGR 2.0 genes in this pathway |
| DER IFN BETA RESPONSE UP | 102 | 67 | All SZGR 2.0 genes in this pathway |
| BROCKE APOPTOSIS REVERSED BY IL6 | 144 | 98 | All SZGR 2.0 genes in this pathway |
| TENEDINI MEGAKARYOCYTE MARKERS | 66 | 48 | All SZGR 2.0 genes in this pathway |
| SASAKI ADULT T CELL LEUKEMIA | 176 | 122 | All SZGR 2.0 genes in this pathway |
| PENG GLUTAMINE DEPRIVATION DN | 337 | 230 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| DER IFN GAMMA RESPONSE UP | 71 | 45 | All SZGR 2.0 genes in this pathway |
| XU RESPONSE TO TRETINOIN AND NSC682994 UP | 17 | 13 | All SZGR 2.0 genes in this pathway |
| NEMETH INFLAMMATORY RESPONSE LPS UP | 88 | 64 | All SZGR 2.0 genes in this pathway |
| BENNETT SYSTEMIC LUPUS ERYTHEMATOSUS | 31 | 21 | All SZGR 2.0 genes in this pathway |
| BASSO CD40 SIGNALING UP | 101 | 76 | All SZGR 2.0 genes in this pathway |
| LIANG SILENCED BY METHYLATION 2 | 53 | 34 | All SZGR 2.0 genes in this pathway |
| TRAYNOR RETT SYNDROM UP | 45 | 33 | All SZGR 2.0 genes in this pathway |
| NATSUME RESPONSE TO INTERFERON BETA UP | 71 | 49 | All SZGR 2.0 genes in this pathway |
| RUAN RESPONSE TO TROGLITAZONE UP | 25 | 15 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION UP | 314 | 201 | All SZGR 2.0 genes in this pathway |
| RUAN RESPONSE TO TNF UP | 12 | 10 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| CHANG IMMORTALIZED BY HPV31 DN | 65 | 45 | All SZGR 2.0 genes in this pathway |
| BROWNE INTERFERON RESPONSIVE GENES | 68 | 44 | All SZGR 2.0 genes in this pathway |
| KRASNOSELSKAYA ILF3 TARGETS UP | 38 | 24 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC DN | 228 | 146 | All SZGR 2.0 genes in this pathway |
| CHIBA RESPONSE TO TSA | 50 | 31 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| RUAN RESPONSE TO TNF TROGLITAZONE UP | 17 | 11 | All SZGR 2.0 genes in this pathway |
| ZHANG ANTIVIRAL RESPONSE TO RIBAVIRIN UP | 30 | 21 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS UP | 198 | 132 | All SZGR 2.0 genes in this pathway |
| LIANG SILENCED BY METHYLATION UP | 32 | 18 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C5 | 46 | 36 | All SZGR 2.0 genes in this pathway |
| MAHAJAN RESPONSE TO IL1A UP | 81 | 52 | All SZGR 2.0 genes in this pathway |
| GERHOLD ADIPOGENESIS UP | 49 | 40 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS DN | 314 | 188 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE DN | 373 | 196 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER T7 | 98 | 63 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| RIGGINS TAMOXIFEN RESISTANCE DN | 220 | 147 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY OVERCONNECTED IN BREAST CANCER | 22 | 19 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE UP | 156 | 92 | All SZGR 2.0 genes in this pathway |
| HELLER SILENCED BY METHYLATION UP | 282 | 183 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS UP | 317 | 208 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION UP | 461 | 298 | All SZGR 2.0 genes in this pathway |
| WALLACE PROSTATE CANCER RACE UP | 299 | 167 | All SZGR 2.0 genes in this pathway |
| OUYANG PROSTATE CANCER PROGRESSION UP | 20 | 14 | All SZGR 2.0 genes in this pathway |
| MOSERLE IFNA RESPONSE | 31 | 19 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP | 374 | 247 | All SZGR 2.0 genes in this pathway |
| ICHIBA GRAFT VERSUS HOST DISEASE D7 UP | 107 | 67 | All SZGR 2.0 genes in this pathway |
| ICHIBA GRAFT VERSUS HOST DISEASE 35D UP | 131 | 79 | All SZGR 2.0 genes in this pathway |
| LINDSTEDT DENDRITIC CELL MATURATION A | 67 | 52 | All SZGR 2.0 genes in this pathway |
| NUTT GBM VS AO GLIOMA UP | 46 | 34 | All SZGR 2.0 genes in this pathway |
| ZHANG INTERFERON RESPONSE | 23 | 14 | All SZGR 2.0 genes in this pathway |
| WUNDER INFLAMMATORY RESPONSE AND CHOLESTEROL UP | 58 | 38 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 DN | 315 | 201 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF UP | 418 | 282 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| COLINA TARGETS OF 4EBP1 AND 4EBP2 | 356 | 214 | All SZGR 2.0 genes in this pathway |
| LIU VAV3 PROSTATE CARCINOGENESIS UP | 89 | 61 | All SZGR 2.0 genes in this pathway |
| JISON SICKLE CELL DISEASE UP | 181 | 106 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G5 DN | 27 | 17 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS INTERFERON UP | 26 | 10 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER LATE RECURRENCE UP | 62 | 42 | All SZGR 2.0 genes in this pathway |
| TSAI DNAJB4 TARGETS UP | 13 | 8 | All SZGR 2.0 genes in this pathway |
| UROSEVIC RESPONSE TO IMIQUIMOD | 23 | 14 | All SZGR 2.0 genes in this pathway |
| HAHTOLA CTCL CUTANEOUS | 26 | 19 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA LB DN | 44 | 23 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO GONADOTROPHINS DN | 87 | 66 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO FORSKOLIN DN | 88 | 68 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE G2 | 182 | 102 | All SZGR 2.0 genes in this pathway |
| STAMBOLSKY TARGETS OF MUTATED TP53 DN | 50 | 24 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 7 | 403 | 240 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 10HR DN | 244 | 157 | All SZGR 2.0 genes in this pathway |
| KRIEG KDM3A TARGETS NOT HYPOXIA | 208 | 107 | All SZGR 2.0 genes in this pathway |
| ALTEMEIER RESPONSE TO LPS WITH MECHANICAL VENTILATION | 128 | 81 | All SZGR 2.0 genes in this pathway |
| DURAND STROMA S UP | 297 | 194 | All SZGR 2.0 genes in this pathway |
| HECKER IFNB1 TARGETS | 95 | 54 | All SZGR 2.0 genes in this pathway |