Gene Page: STX5
Summary ?
| GeneID | 6811 |
| Symbol | STX5 |
| Synonyms | SED5|STX5A |
| Description | syntaxin 5 |
| Reference | MIM:603189|HGNC:HGNC:11440|Ensembl:ENSG00000162236|HPRD:04426|Vega:OTTHUMG00000143864 |
| Gene type | protein-coding |
| Map location | 11q12.3 |
| Pascal p-value | 0.146 |
| Sherlock p-value | 0.853 |
| Fetal beta | 0.501 |
| DMG | 1 (# studies) |
| Support | INTRACELLULAR TRAFFICKING |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 1 |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg14519525 | 11 | 62414275 | STX5 | 1.46E-5 | 4.063 | DMG:vanEijk_2014 |
Section II. Transcriptome annotation
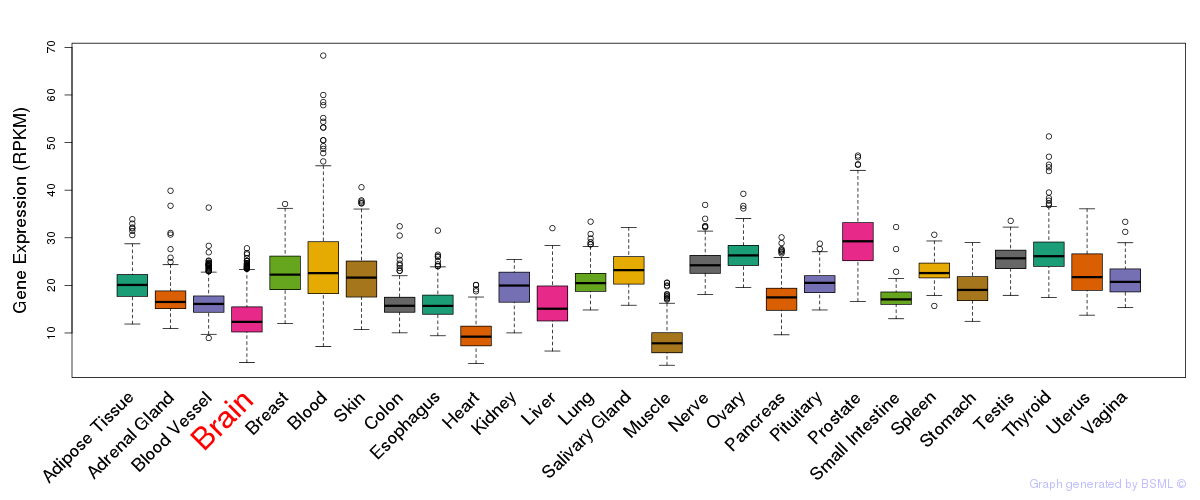
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005484 | SNAP receptor activity | IDA | 15215310 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006903 | vesicle targeting | TAS | 9188044 | |
| GO:0006886 | intracellular protein transport | IEA | - | |
| GO:0042147 | retrograde transport, endosome to Golgi | IDA | 15215310 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0031201 | SNARE complex | TAS | neuron, Synap (GO term level: 6) | 15215310 |
| GO:0000139 | Golgi membrane | IEA | - | |
| GO:0005794 | Golgi apparatus | IDA | 15215310 | |
| GO:0005634 | nucleus | IDA | 18029348 | |
| GO:0005730 | nucleolus | IDA | 18029348 | |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0033116 | ER-Golgi intermediate compartment membrane | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| APLP1 | APLP | amyloid beta (A4) precursor-like protein 1 | Two-hybrid | BioGRID | 16169070 |
| BAT3 | BAG-6 | BAG6 | D6S52E | G3 | HLA-B associated transcript 3 | Two-hybrid | BioGRID | 16169070 |
| BET1 | DKFZp781C0425 | HBET1 | blocked early in transport 1 homolog (S. cerevisiae) | - | HPRD | 9094723 |
| BET1 | DKFZp781C0425 | HBET1 | blocked early in transport 1 homolog (S. cerevisiae) | Affinity Capture-Western | BioGRID | 11927603 |
| BET1L | BET1L1 | GOLIM3 | GS15 | HSPC197 | blocked early in transport 1 homolog (S. cerevisiae)-like | Affinity Capture-Western | BioGRID | 11927603 |12388752 |
| C1orf103 | FLJ11269 | RIF1 | RP11-96K19.1 | chromosome 1 open reading frame 103 | Two-hybrid | BioGRID | 16169070 |
| COPS6 | CSN6 | MOV34-34KD | COP9 constitutive photomorphogenic homolog subunit 6 (Arabidopsis) | Two-hybrid | BioGRID | 16169070 |
| GOSR1 | GOLIM2 | GOS-28 | GOS28 | GOS28/P28 | GS28 | P28 | golgi SNAP receptor complex member 1 | - | HPRD,BioGRID | 9094723 |
| GOSR2 | Bos1 | GS27 | golgi SNAP receptor complex member 2 | Affinity Capture-Western Co-purification Two-hybrid | BioGRID | 9094723 |9647643 |11927603 |
| GOSR2 | Bos1 | GS27 | golgi SNAP receptor complex member 2 | - | HPRD | 9094723 |9647643|11035026 |
| HAP1 | HAP2 | HIP5 | HLP | hHLP1 | huntingtin-associated protein 1 | Two-hybrid | BioGRID | 16169070 |
| IMMT | DKFZp779P1653 | HMP | MGC111146 | P87 | P87/89 | P89 | PIG4 | PIG52 | inner membrane protein, mitochondrial (mitofilin) | Two-hybrid | BioGRID | 16169070 |
| NAPA | SNAPA | N-ethylmaleimide-sensitive factor attachment protein, alpha | Reconstituted Complex Two-hybrid | BioGRID | 9506515 |16189514 |
| NSFL1C | MGC3347 | UBX1 | UBXD10 | UBXN2C | dJ776F14.1 | p47 | NSFL1 (p97) cofactor (p47) | - | HPRD | 9506515 |
| NSFL1C | MGC3347 | UBX1 | UBXD10 | UBXN2C | dJ776F14.1 | p47 | NSFL1 (p97) cofactor (p47) | - | HPRD,BioGRID | 9506515|12473691 |
| PSEN1 | AD3 | FAD | PS1 | S182 | presenilin 1 | The full-length PS1 holoprotein interacts with Syntaxin 5. | BIND | 15109302 |
| SCFD1 | C14orf163 | KIAA0917 | RA410 | SLY1 | STXBP1L2 | sec1 family domain containing 1 | - | HPRD | 12506202 |14565970 |
| SEC22A | SEC22L2 | SEC22 vesicle trafficking protein homolog A (S. cerevisiae) | Affinity Capture-Western | BioGRID | 11927603 |
| SEC22B | ERS-24 | SEC22L1 | SEC22 vesicle trafficking protein homolog B (S. cerevisiae) | - | HPRD | 11035026 |
| SEC22B | ERS-24 | SEC22L1 | SEC22 vesicle trafficking protein homolog B (S. cerevisiae) | Affinity Capture-Western | BioGRID | 15029241 |
| SNAP23 | HsT17016 | SNAP23A | SNAP23B | synaptosomal-associated protein, 23kDa | Reconstituted Complex | BioGRID | 12651853 |
| SNAP25 | FLJ23079 | RIC-4 | RIC4 | SEC9 | SNAP | SNAP-25 | bA416N4.2 | dJ1068F16.2 | synaptosomal-associated protein, 25kDa | Reconstituted Complex | BioGRID | 9325254 |
| STX17 | FLJ20651 | MGC102796 | MGC126613 | MGC126615 | syntaxin 17 | Co-purification | BioGRID | 9094723 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | Two-hybrid | BioGRID | 16169070 |
| USO1 | P115 | TAP | VDP | USO1 homolog, vesicle docking protein (yeast) | - | HPRD,BioGRID | 11927603 |
| VAPB | ALS8 | VAMP-B | VAMP-C | VAP-B | VAP-C | VAMP (vesicle-associated membrane protein)-associated protein B and C | Reconstituted Complex | BioGRID | 12651853 |
| VCP | IBMPFD | MGC131997 | MGC148092 | MGC8560 | TERA | p97 | valosin-containing protein | - | HPRD | 9506515 |
| VCPIP1 | DKFZp686G038 | DUBA3 | FLJ23132 | FLJ60694 | KIAA1850 | VCIP135 | valosin containing protein (p97)/p47 complex interacting protein 1 | - | HPRD | 12473691 |
| VTI1B | VTI1 | VTI1-LIKE | VTI1L | VTI2 | vesicle transport through interaction with t-SNAREs homolog 1B (yeast) | Affinity Capture-Western | BioGRID | 11927603 |
| YKT6 | - | YKT6 v-SNARE homolog (S. cerevisiae) | Affinity Capture-Western | BioGRID | 11927603 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG SNARE INTERACTIONS IN VESICULAR TRANSPORT | 38 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME BOTULINUM NEUROTOXICITY | 19 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | 17 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR DN | 214 | 133 | All SZGR 2.0 genes in this pathway |
| OUELLET OVARIAN CANCER INVASIVE VS LMP DN | 11 | 8 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| TANAKA METHYLATED IN ESOPHAGEAL CARCINOMA | 103 | 58 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| FERREIRA EWINGS SARCOMA UNSTABLE VS STABLE DN | 98 | 59 | All SZGR 2.0 genes in this pathway |
| CAFFAREL RESPONSE TO THC 8HR 5 UP | 16 | 7 | All SZGR 2.0 genes in this pathway |
| SAKAI TUMOR INFILTRATING MONOCYTES DN | 81 | 51 | All SZGR 2.0 genes in this pathway |
| UEDA CENTRAL CLOCK | 88 | 62 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN UP | 439 | 257 | All SZGR 2.0 genes in this pathway |
| MALONEY RESPONSE TO 17AAG UP | 41 | 26 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB UP | 245 | 159 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR UP | 293 | 203 | All SZGR 2.0 genes in this pathway |
| COULOUARN TEMPORAL TGFB1 SIGNATURE DN | 138 | 99 | All SZGR 2.0 genes in this pathway |
| KYNG WERNER SYNDROM AND NORMAL AGING DN | 225 | 124 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-133 | 404 | 411 | 1A,m8 | hsa-miR-133a | UUGGUCCCCUUCAACCAGCUGU |
| hsa-miR-133b | UUGGUCCCCUUCAACCAGCUA | ||||
| miR-150 | 91 | 98 | 1A,m8 | hsa-miR-150 | UCUCCCAACCCUUGUACCAGUG |
| miR-182 | 518 | 524 | 1A | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| miR-24 | 72 | 78 | m8 | hsa-miR-24SZ | UGGCUCAGUUCAGCAGGAACAG |
| hsa-miR-24SZ | UGGCUCAGUUCAGCAGGAACAG | ||||
| miR-27 | 232 | 238 | 1A | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-431 | 555 | 561 | m8 | hsa-miR-431 | UGUCUUGCAGGCCGUCAUGCA |
| miR-485-5p | 565 | 571 | m8 | hsa-miR-485-5p | AGAGGCUGGCCGUGAUGAAUUC |
| miR-96 | 518 | 524 | 1A | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.