Gene Page: HNF1A
Summary ?
| GeneID | 6927 |
| Symbol | HNF1A |
| Synonyms | HNF-1A|HNF1|IDDM20|LFB1|MODY3|TCF-1|TCF1 |
| Description | HNF1 homeobox A |
| Reference | MIM:142410|HGNC:HGNC:11621|Ensembl:ENSG00000135100|HPRD:00800|Vega:OTTHUMG00000151015 |
| Gene type | protein-coding |
| Map location | 12q24.2 |
| Pascal p-value | 0.007 |
| Fetal beta | -0.091 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0538 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg01341572 | 12 | 121416512 | HNF1A | 3.161E-4 | 0.343 | 0.04 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
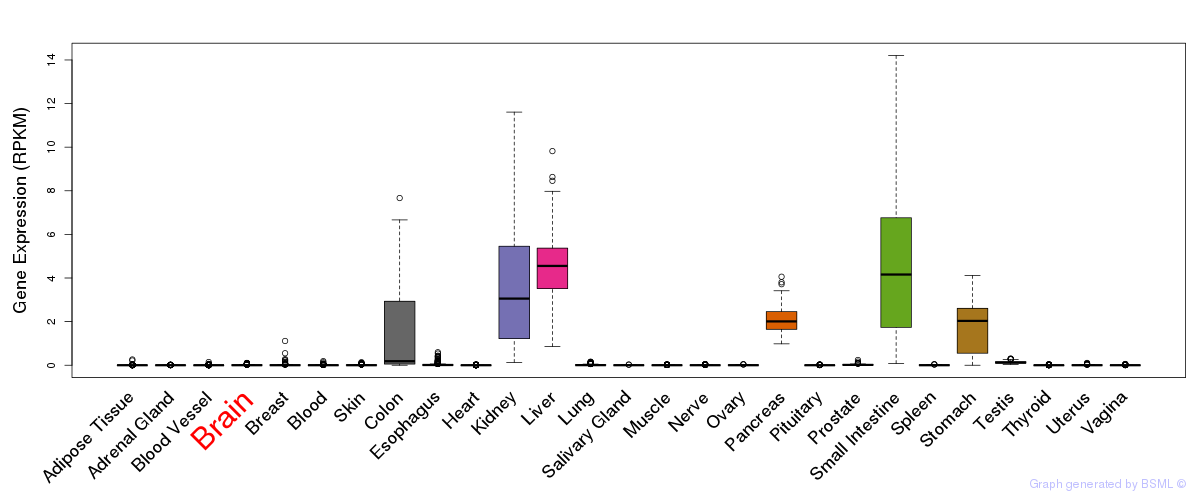
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003700 | transcription factor activity | IDA | 11980910 | |
| GO:0003702 | RNA polymerase II transcription factor activity | IDA | 10330009 | |
| GO:0005515 | protein binding | IPI | 11980910 | |
| GO:0016563 | transcription activator activity | IEA | - | |
| GO:0043565 | sequence-specific DNA binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0010551 | regulation of specific transcription from RNA polymerase II promoter | IDA | 10330009 | |
| GO:0042593 | glucose homeostasis | IMP | 15277395 | |
| GO:0045893 | positive regulation of transcription, DNA-dependent | IDA | 11980910 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0043234 | protein complex | IDA | 11980910 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AES | AES-1 | AES-2 | ESP1 | GRG | GRG5 | TLE5 | amino-terminal enhancer of split | Two-hybrid | BioGRID | 11266540 |
| ALB | DKFZp779N1935 | PRO0883 | PRO0903 | PRO1341 | albumin | - | HPRD | 2460858 |
| CALM3 | PHKD | PHKD3 | calmodulin 3 (phosphorylase kinase, delta) | Affinity Capture-MS | BioGRID | 17353931 |
| CDX2 | CDX-3 | CDX3 | caudal type homeobox 2 | - | HPRD,BioGRID | 10677375 |
| CEBPA | C/EBP-alpha | CEBP | CCAAT/enhancer binding protein (C/EBP), alpha | - | HPRD | 8288579 |
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | CBP interacts with the HNF-1 gene. | BIND | 15616580 |
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | - | HPRD,BioGRID | 10777539 |
| CTNNB1 | CTNNB | DKFZp686D02253 | FLJ25606 | FLJ37923 | catenin (cadherin-associated protein), beta 1, 88kDa | - | HPRD | 11266540 |12107263 |
| ELP3 | FLJ10422 | KAT9 | elongation protein 3 homolog (S. cerevisiae) | Elp3 interacts with the HNF-1 gene. | BIND | 15616580 |
| EP300 | KAT3B | p300 | E1A binding protein p300 | - | HPRD,BioGRID | 11978637 |
| FOS | AP-1 | C-FOS | v-fos FBJ murine osteosarcoma viral oncogene homolog | - | HPRD,BioGRID | 11134330 |
| GATA5 | bB379O24.1 | GATA binding protein 5 | - | HPRD | 12011060 |14715527 |
| H3F3A | H3.3A | H3F3 | MGC87782 | MGC87783 | H3 histone, family 3A | H3F3A (Histone 3) interacts with the TCF1 (HNF-1) gene. | BIND | 15616580 |
| HIST4H4 | H4/p | MGC24116 | histone cluster 4, H4 | HIST4H4 (Histone 4) interacts with the TCF1 (HNF-1) gene. | BIND | 15616580 |
| HNF1A | HNF1 | LFB1 | MODY3 | TCF1 | HNF1 homeobox A | - | HPRD,BioGRID | 2044952 |
| HNF1B | FJHN | HNF1beta | HNF2 | HPC11 | LF-B3 | LFB3 | MODY5 | TCF2 | VHNF1 | HNF1 homeobox B | Reconstituted Complex | BioGRID | 2044952 |
| HNF4A | FLJ39654 | HNF4 | HNF4a7 | HNF4a8 | HNF4a9 | MODY | MODY1 | NR2A1 | NR2A21 | TCF | TCF14 | hepatocyte nuclear factor 4, alpha | - | HPRD | 9792724 |
| KAT2B | CAF | P | P/CAF | PCAF | K(lysine) acetyltransferase 2B | - | HPRD,BioGRID | 10777539 |
| PCBD1 | DCOH | PCBD | PCD | PHS | pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha | - | HPRD,BioGRID | 9092652 |
| PCBD2 | DCOH2 | DCOHM | PHS2 | pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha (TCF1) 2 | Affinity Capture-MS | BioGRID | 17353931 |
| RAC3 | - | ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) | - | HPRD,BioGRID | 10777539 |
| RHPN2 | RhoBP | p76RBE | rhophilin, Rho GTPase binding protein 2 | Affinity Capture-MS | BioGRID | 17353931 |
| SMARCA4 | BAF190 | BRG1 | FLJ39786 | SNF2 | SNF2-BETA | SNF2L4 | SNF2LB | SWI2 | hSNF2b | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 | Brg-1 interacts with the HNF-1 gene. | BIND | 15616580 |
| SMARCA5 | ISWI | SNF2H | WCRF135 | hISWI | hSNF2H | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 | SNF2H interacts with the HNF-1 gene. | BIND | 15616580 |
| SNRPA1 | - | small nuclear ribonucleoprotein polypeptide A' | Affinity Capture-MS | BioGRID | 17353931 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | - | HPRD,BioGRID | 10777539 |
| SSRP1 | FACT | FACT80 | T160 | structure specific recognition protein 1 | FACT interacts with the HNF-1 gene. | BIND | 15616580 |
| STAT3 | APRF | FLJ20882 | HIES | MGC16063 | signal transducer and activator of transcription 3 (acute-phase response factor) | - | HPRD,BioGRID | 11134330 |
| TLE1 | ESG | ESG1 | GRG1 | transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila) | - | HPRD,BioGRID | 10209158 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MATURITY ONSET DIABETES OF THE YOUNG | 25 | 19 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ALK PATHWAY | 37 | 29 | All SZGR 2.0 genes in this pathway |
| BIOCARTA GH PATHWAY | 28 | 22 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PS1 PATHWAY | 14 | 14 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MAL PATHWAY | 19 | 17 | All SZGR 2.0 genes in this pathway |
| PID PS1 PATHWAY | 46 | 39 | All SZGR 2.0 genes in this pathway |
| PID HNF3B PATHWAY | 45 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF BETA CELL DEVELOPMENT | 30 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | 20 | 15 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| MA MYELOID DIFFERENTIATION DN | 44 | 30 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN MGMT 48HR DN | 161 | 105 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY OVERCONNECTED IN BREAST CANCER | 22 | 19 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS UP | 170 | 107 | All SZGR 2.0 genes in this pathway |
| MATZUK IMPLANTATION AND UTERINE | 22 | 15 | All SZGR 2.0 genes in this pathway |
| ZHOU PANCREATIC BETA CELL | 11 | 9 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| GU PDEF TARGETS DN | 39 | 20 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 | 718 | 401 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 2 | 86 | 50 | All SZGR 2.0 genes in this pathway |